

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YPL072W | OTHER | 0.868898 | 0.116899 | 0.014203 |
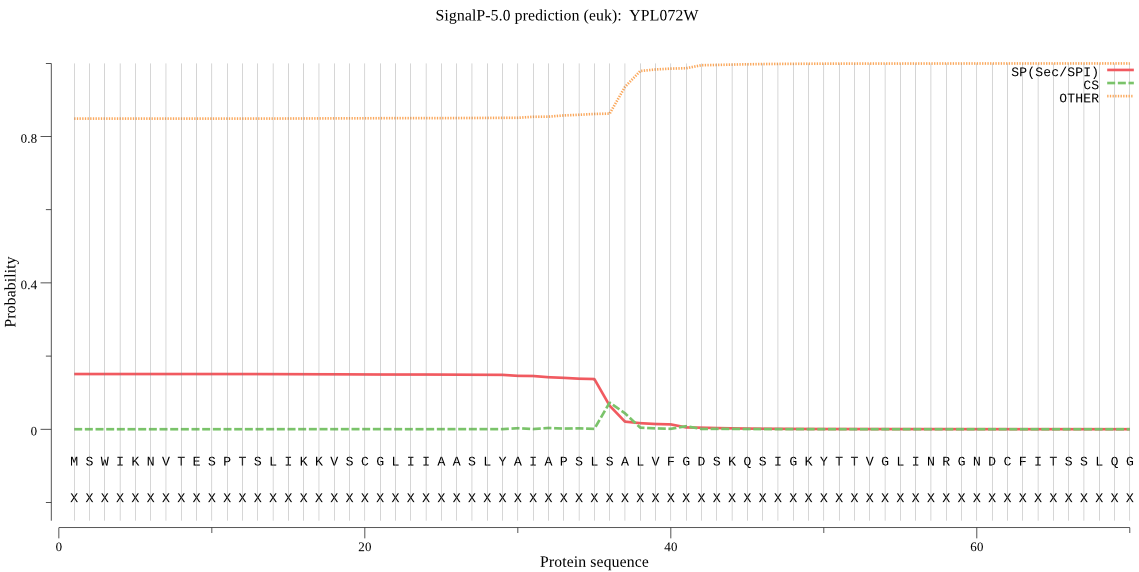
MSWIKNVTESPTSLIKKVSCGLIIAASLYAIAPSLSALVFGDSKQSIGKYTTVGLINRGN DCFITSSLQGLAGIPRFVEYLKRIRTVLLELETKLSNNAKGDNPTVDNTTRHSRLENSSN SLAPLHESLTSLILDLISVKDRKTSISPKIVINTLESIFKSKISSKQNDAHEFTLILLQT LQEERSKLIDYSKQICNLNIPKFPFEGETSKFLVCLKCKGLSEPSYKQTFIRELSVPQQT SENLSNILAHDETEIIDDYSCLICQIRAILNHEEYRNFKDCTPDEILMLDRLKNYATKAP INENLPFEVEQYVKRYSKGNLQVSNIKGKVIKKDVVVQLPDILIVHLSRSTFNGITYSRN PCNVKFGERITLSEYTLAESGTITENRQVKYNLKSVVKHTGSHSSGHYMCYRRKTEIRFG KEDESSFRRAPVVNNEVNKNREQNVAHNDYKKSRYKKVKNALRYPYWQISDTAIKESTAS TVLNEQKYAYMLYYERVNK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YPL072W | 164 S | KSKISSKQN | 0.995 | unsp | YPL072W | 164 S | KSKISSKQN | 0.995 | unsp | YPL072W | 164 S | KSKISSKQN | 0.995 | unsp | YPL072W | 225 S | LSEPSYKQT | 0.993 | unsp | YPL072W | 426 S | EDESSFRRA | 0.992 | unsp | YPL072W | 113 S | TTRHSRLEN | 0.995 | unsp | YPL072W | 138 S | LDLISVKDR | 0.996 | unsp |