

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
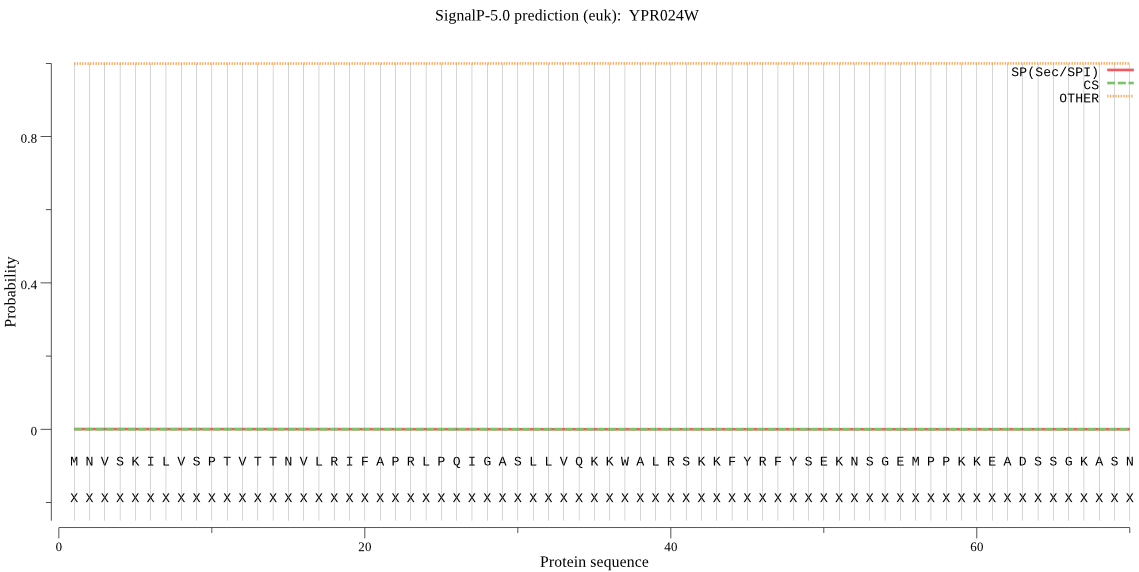
| YPR024W | OTHER | 0.791669 | 0.000294 | 0.208038 |
MNVSKILVSPTVTTNVLRIFAPRLPQIGASLLVQKKWALRSKKFYRFYSEKNSGEMPPKK EADSSGKASNKSTISSIDNSQPPPPSNTNDKTKQANVAVSHAMLATREQEANKDLTSPDA QAAFYKLLLQSNYPQYVVSRFETPGIASSPECMELYMEALQRIGRHSEADAVRQNLLTAS SAGAVNPSLASSSSNQSGYHGNFPSMYSPLYGSRKEPLHVVVSESTFTVVSRWVKWLLVF GILTYSFSEGFKYITENTTLLKSSEVADKSVDVAKTNVKFDDVCGCDEARAELEEIVDFL KDPTKYESLGGKLPKGVLLTGPPGTGKTLLARATAGEAGVDFFFMSGSEFDEVYVGVGAK RIRDLFAQARSRAPAIIFIDELDAIGGKRNPKDQAYAKQTLNQLLVELDGFSQTSGIIII GATNFPEALDKALTRPGRFDKVVNVDLPDVRGRADILKHHMKKITLADNVDPTIIARGTP GLSGAELANLVNQAAVYACQKNAVSVDMSHFEWAKDKILMGAERKTMVLTDAARKATAFH EAGHAIMAKYTNGATPLYKATILPRGRALGITFQLPEMDKVDITKRECQARLDVCMGGKI AEELIYGKDNTTSGCGSDLQSATGTARAMVTQYGMSDDVGPVNLSENWESWSNKIRDIAD NEVIELLKDSEERARRLLTKKNVELHRLAQGLIEYETLDAHEIEQVCKGEKLDKLKTSTN TVVEGPDSDERKDIGDDKPKIPTMLNA
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| YPR024W | T | 73 | 0.551 | 0.023 | YPR024W | T | 88 | 0.500 | 0.042 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| YPR024W | T | 73 | 0.551 | 0.023 | YPR024W | T | 88 | 0.500 | 0.042 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YPR024W | 69 S | SGKASNKST | 0.992 | unsp | YPR024W | 69 S | SGKASNKST | 0.992 | unsp | YPR024W | 69 S | SGKASNKST | 0.992 | unsp | YPR024W | 167 S | IGRHSEADA | 0.998 | unsp | YPR024W | 213 S | PLYGSRKEP | 0.993 | unsp | YPR024W | 263 S | TLLKSSEVA | 0.99 | unsp | YPR024W | 670 S | LLKDSEERA | 0.995 | unsp | YPR024W | 728 S | EGPDSDERK | 0.995 | unsp | YPR024W | 49 S | YRFYSEKNS | 0.992 | unsp | YPR024W | 53 S | SEKNSGEMP | 0.991 | unsp |