

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
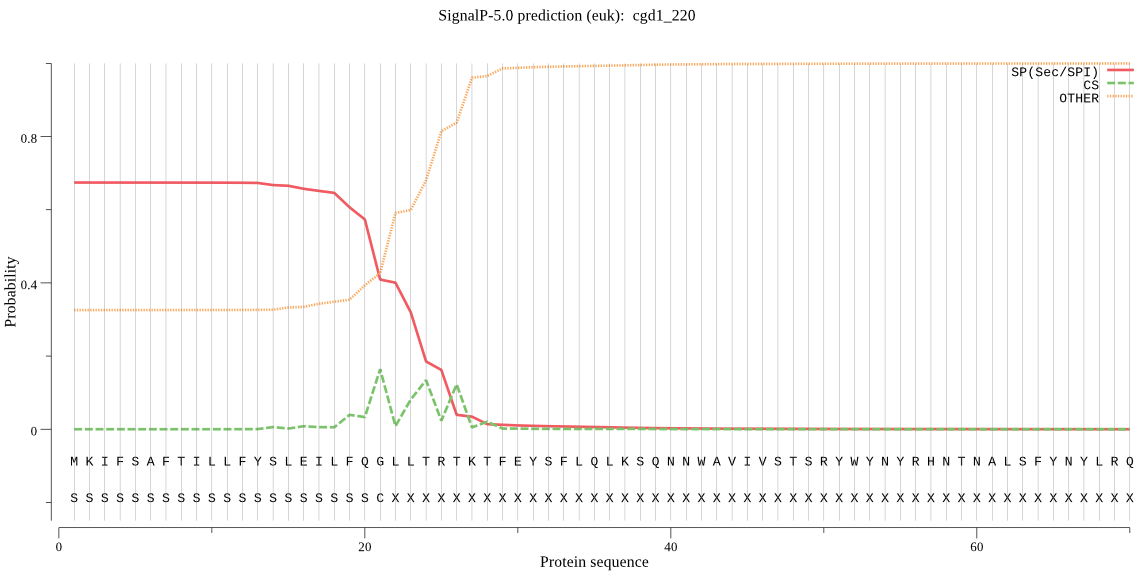
| cgd1_220 | SP | 0.139417 | 0.860219 | 0.000364 | CS pos: 24-25. LLT-RT. Pr: 0.2635 |
MKIFSAFTILLFYSLEILFQGLLTRTKTFEYSFLQLKSQNNWAVIVSTSRYWYNYRHNTN ALSFYNYLRQNGFRDDRIILMLAENIPCNTRNSIPGGVYSEDFDFFYNLNNHTQTMECAD VDYREDEVTVSNFIKVLTNKHDDSVPNKKRLLSDEDSNIFIFLTGHGGDGFLKFQDFEEM TSFELANAIKEMKAHKRFKKMFIISETCQASTLHNHLDFEDVYAIGCSSLGESSYSKHYK VEIGVASIDRFTHFSLADFKNLNRNKLMPIASLIGKYSVFQLKSTPQLKYKPGKTDIKNV YVNEFFFPNIEKLFSLNIGNLILNHKNIKEPIKQVLNEGASKYLNCSLLYHTLKRYNDLD LIDSLSNCHFKNTLKKAIIFSQKRYVFNSKFSSKKMIDNKSLMLIKAIFGLTFILILVFI LSYYSL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| cgd1_220 | 153 S | KRLLSDEDS | 0.998 | unsp |