

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
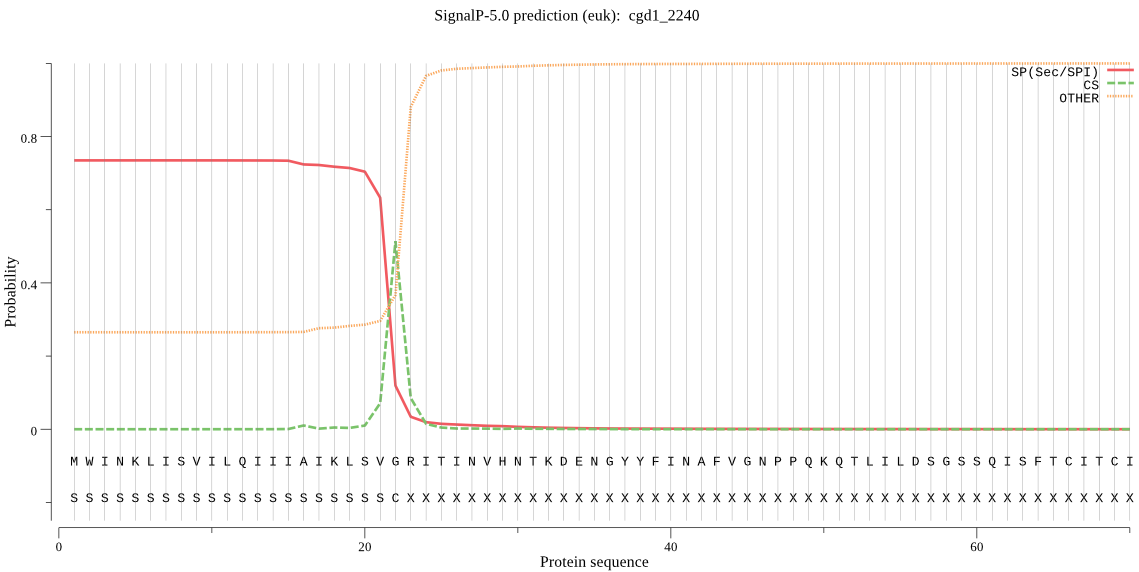
| cgd1_2240 | SP | 0.029357 | 0.969226 | 0.001418 | CS pos: 22-23. SVG-RI. Pr: 0.9311 |
MWINKLISVILQIIIAIKLSVGRITINVHNTKDENGYYFINAFVGNPPQKQTLILDSGSS QISFTCITCINCGSHEYPPFDIMKSITGKNCNRKLLGGEKCKYFHRFNEGSVISGKYFSD TLRFEEVQQNSGIIKDHFEIKYDYLGCNELETKKIYRQRATGVFGIGLKSSLDDHVNIIN SLLTSLESNTNIKGSNNLVISICLLYSGGRIVIGEQDEKITNRNSNFSENKRNHVYWVPI IYPSNVYKVSLEGLSIGSGKFSLLEEETSLFAIVDVGSTYSFFPSNLYNKIINKFSKLCE LLNKLDINKCITINQSLCFSDPAKLHALLPIMNIKFGGQPNLIKWIHSSYLIKRERAWCV GIKEQTSYQNHIILGVSFMKKRQIILDPRKKRIGFNLNTTARCKYEQNKTMTNQVKLLNE YTRSK