

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd1_2490 | OTHER | 0.999856 | 0.000069 | 0.000076 |
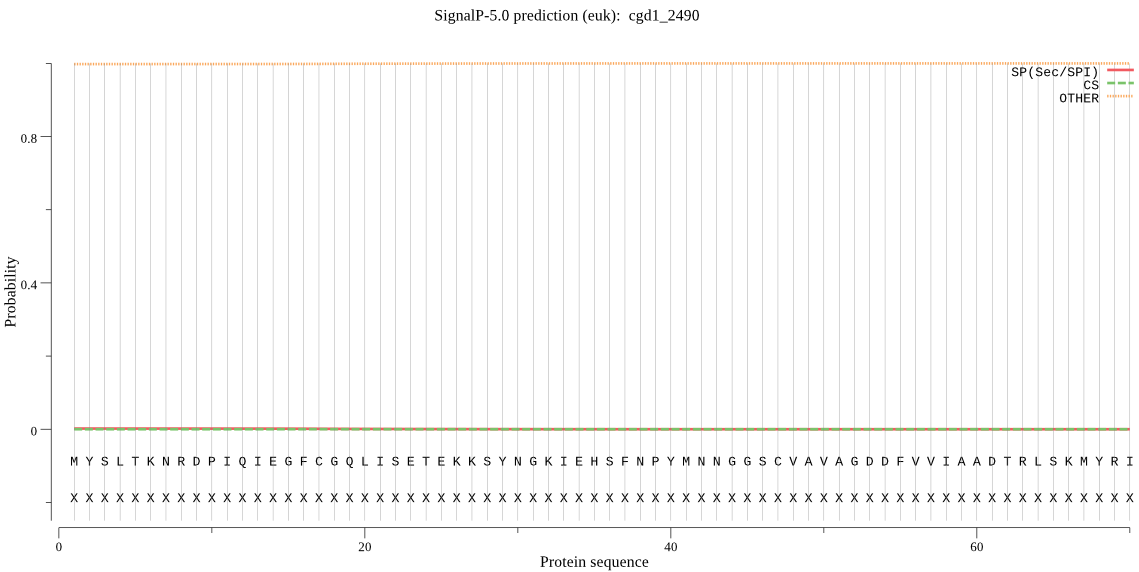
MYSLTKNRDPIQIEGFCGQLISETEKKSYNGKIEHSFNPYMNNGGSCVAVAGDDFVVIAA DTRLSKMYRIASRSVSKTCQLTNKCILACSGMLADINALRKTLLAKVKLYEFEHNKTPSI NAIAQLLSCILYSKRFFPYYSFCLLSGLDEQGKGVVYGYDAVGSFDQHKFVALGSGGSLI TSILDNQISGNNQTSFESIDKVGIINIVKDSITSASERDVHTGDSAEVIVIDSSGISVSS LRLRED
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| cgd1_2490 | 76 S | SRSVSKTCQ | 0.993 | unsp | cgd1_2490 | 216 S | ITSASERDV | 0.993 | unsp |