

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd1_2870 | SP | 0.010906 | 0.988679 | 0.000415 | CS pos: 24-25. CHG-NK. Pr: 0.8819 |
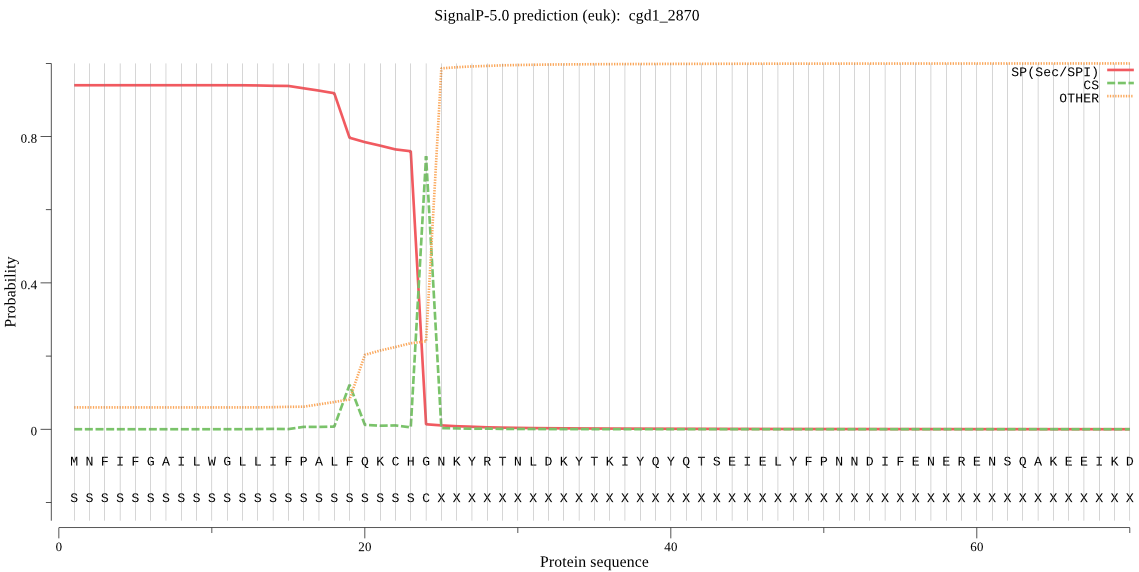
MNFIFGAILWGLLIFPALFQKCHGNKYRTNLDKYTKIYQYQTSEIELYFPNNDIFENERE NSQAKEEIKDLKSTKVSDIRYSFMKLGRKEFLCGFEGYGKNDSLNLSEETNQSWDNMNSK VKNYLKKSKISWLMERCFIYTKKKDINNGIHVQVDNFEICVGVSVRHKRQIIDKETSDNI LESQFNIVGDYKPTEDTFYENGTIIQYYNPAIENLKGDSGYSASIEFKCSYNKSGISQVT EDVNKEFKPVIKVDFLSPSFCDWRVDESKNITSLHKLESLLLPLENRCQNFTDSRFWNYE LCNLYVVSQFKKDSSTQELRLFNLGIHPNTAQILNITDDLEEYLSKNLISNIVNTIKPGR VSLLENITTKIEPRNINELISGKYVHSKYVITVMLLNGTYCPENNQKRSIKLVFECPDNF ESISDYFRIVNVIEDSTCNYEMLIISPVICSHPLLMPPPIYKHGRVKCIPKNLIINKLIE DESTEHLFESQETRSESGLVSISQIGNQFEAVNIEYIDSTKNGSSTQHFFTLSDSPRIRS SNLTSPKFLVGQIVQHLWWNYYGVVVGWDWELNAPKQWADNIYQKYPPESRKRPHYLLLV HQEETLFNNSSPIESIPSNLTHTYIPEIALKHVPNLDSDNHFSNKDIIINKYLNTYFSYW SEIHQRFIPNTNSTLWRIYPNDFDKLNERDEL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd1_2870 | 545 S | SNLTSPKFL | 0.992 | unsp | cgd1_2870 | 545 S | SNLTSPKFL | 0.992 | unsp | cgd1_2870 | 545 S | SNLTSPKFL | 0.992 | unsp | cgd1_2870 | 590 S | YPPESRKRP | 0.997 | unsp | cgd1_2870 | 73 S | KDLKSTKVS | 0.996 | unsp | cgd1_2870 | 315 S | KKDSSTQEL | 0.995 | unsp |