

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
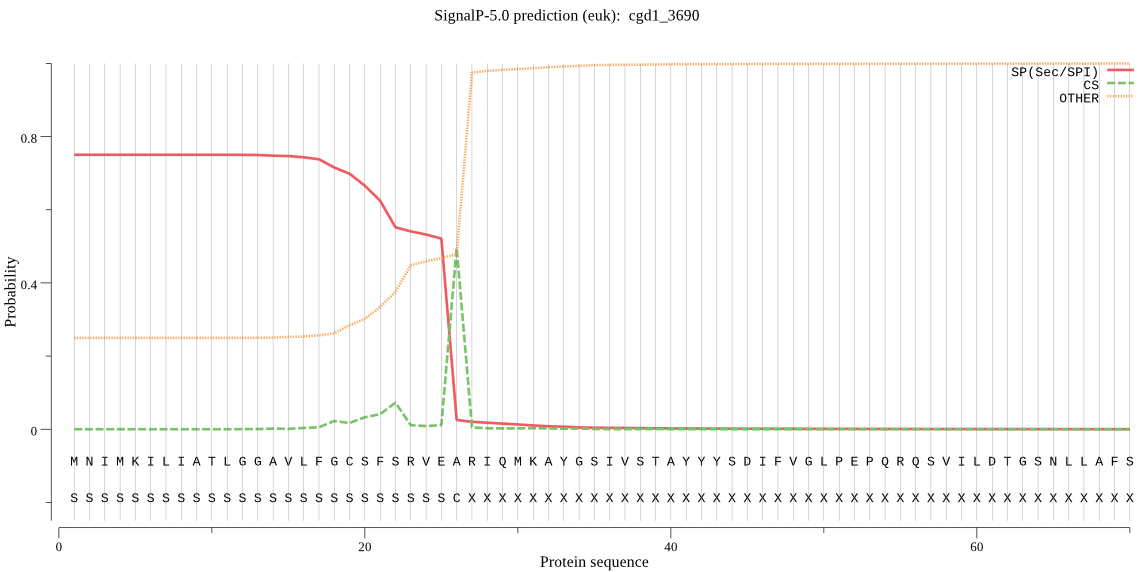
| cgd1_3690 | SP | 0.009233 | 0.990046 | 0.000721 | CS pos: 26-27. VEA-RI. Pr: 0.5338 |
MNIMKILIATLGGAVLFGCSFSRVEARIQMKAYGSIVSTAYYYSDIFVGLPEPQRQSVIL DTGSNLLAFSSTQCQQCGTHLDAYYDPFKSITKREVPCHSYCKVCVNNKKQCAYTIHYLE GSSLSGSYFEDFVAIRNEKGVNSEPSPYVVGLSTIFGGITHETNLFFTQAASGILGLAYT ASSQERAPLFQTWTKRSKYAKDAILSLCFSSEGGMISFGGYNSEYWVLGSNDKSFRSTSE SNLFERALGYSFLSSSSNSQSRSNSRGNNLDSKIGWTPLSILNGNYYVQLTKVSVYQTKL TLHKDSSSGNTKPIPLVIDSGTTLSYFPEHIFIQILNVINQRIAETENKSTFRSLIEAGS KFLEYTGLKSSSSQNELELGIKQISIFPGEIPGAALLRKRVYRRLNNIEDIVNTFNSTVH PENFTNEDIESSNNQLNNSNEVSNHFNQTTTSTTTSTINTLFSSQEFDIDSSVSRIMLET SKGERCWKLRDPNEMSRFPTITLGFPGLKVEWEPAQYLYKKYRNTYCLGFDSDKSFLVLG ASFFINKDVIIDVKNSRASFVKSNCPQIAHARRTSTSEMSQLLKDASTSSPESIMKSEVV VFDSNTKHINIENGNSNNNNINNQNPNLSSGTSSKNPPNYRHSADNDTISESHWEPKNLT SPSSLSSYLSPSAVQYTGVTNHSVDYYNQSINYENLGNNDMLILYLKQQLHDIDDIADNT ENTQIQKNVQDANNSPSDSKNENQDNNSDNKNSSSRYSNSQPTIIHWLIILLSALFSCYL FNGVDQKDVKTE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd1_3690 | 575 S | ARRTSTSEM | 0.997 | unsp | cgd1_3690 | 575 S | ARRTSTSEM | 0.997 | unsp | cgd1_3690 | 575 S | ARRTSTSEM | 0.997 | unsp | cgd1_3690 | 590 S | ASTSSPESI | 0.991 | unsp | cgd1_3690 | 593 S | SSPESIMKS | 0.994 | unsp | cgd1_3690 | 633 S | SSGTSSKNP | 0.993 | unsp | cgd1_3690 | 735 S | DANNSPSDS | 0.996 | unsp | cgd1_3690 | 183 S | YTASSQERA | 0.996 | unsp | cgd1_3690 | 239 S | FRSTSESNL | 0.992 | unsp |