

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd1_440 | OTHER | 0.966214 | 0.028643 | 0.005143 |
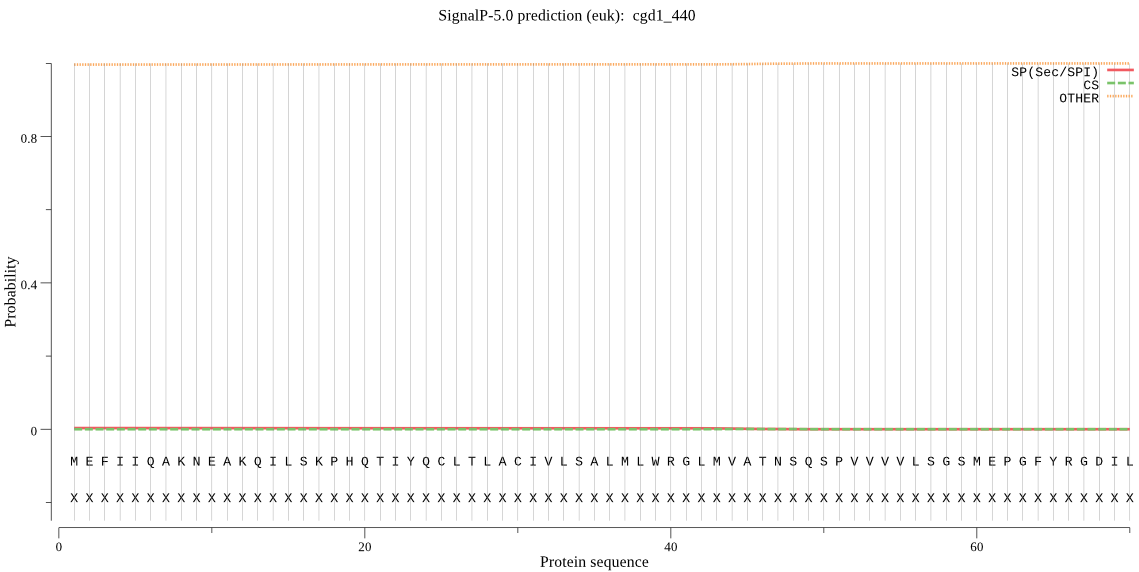
MEFIIQAKNEAKQILSKPHQTIYQCLTLACIVLSALMLWRGLMVATNSQSPVVVVLSGSM EPGFYRGDILFLYNRKSITIGDIVVFSLEGRDIPIVHRVLSYHEGPNDGEISILTKGDNN DVDDRGLYNENQFWLNNKHIMGTAVGIIPKVGMITIWLNDYPWLKYALVGMMGITVLLGK E
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| cgd1_440 | 101 S | HRVLSYHEG | 0.995 | unsp |