

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd2_3660 | SP | 0.026839 | 0.971210 | 0.001951 | CS pos: 20-21. VRS-QN. Pr: 0.9500 |
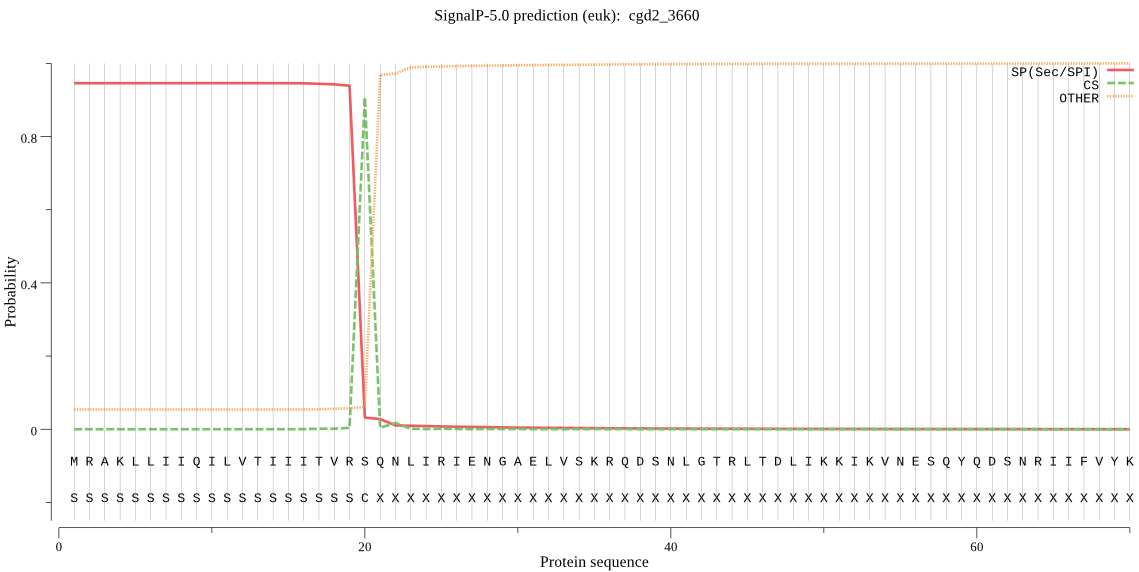
MRAKLLIIQILVTIIITVRSQNLIRIENGAELVSKRQDSNLGTRLTDLIKKIKVNESQYQ DSNRIIFVYKDSNSGSSELFLRLRLRFKNIIERAFNILKFNNTKTRYLKNLSMEIIQTKD NDQMTKFLNVIKDQVSLFQDLIQGVYLDEFIGFEENNYRSTFEIPKINNSIRPTITTENY STIYERQWFHNDSKFGIKSAKMWKRVSELKDQGMGNVVIAVIDSGIDFEHPDLRGKIWRN FGEFDCNDGIDDDMNGYVDDCYGWNFVDNNKIPVDNNGHGTHISGIISAIPNKEVGISGI CWFCQIMVLKVLDSKIRGFLSGFVEAIDYALDKGVRVSNHSYGSRSKSIELLRLAIKRSE DQGMLVIVASGNYESDRNNDIVPTFPSSFKSENIISVTSITSNGELMERATYGRKTVHIG APGVNICSTYLQGEYRCMDGSSFSAPIITGIAGVLLSVFPNISIYSIKDCILKGATRIRG LPG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| cgd2_3660 | 39 S | KRQDSNLGT | 0.993 | unsp | cgd2_3660 | 348 S | SRSKSIELL | 0.996 | unsp |