

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
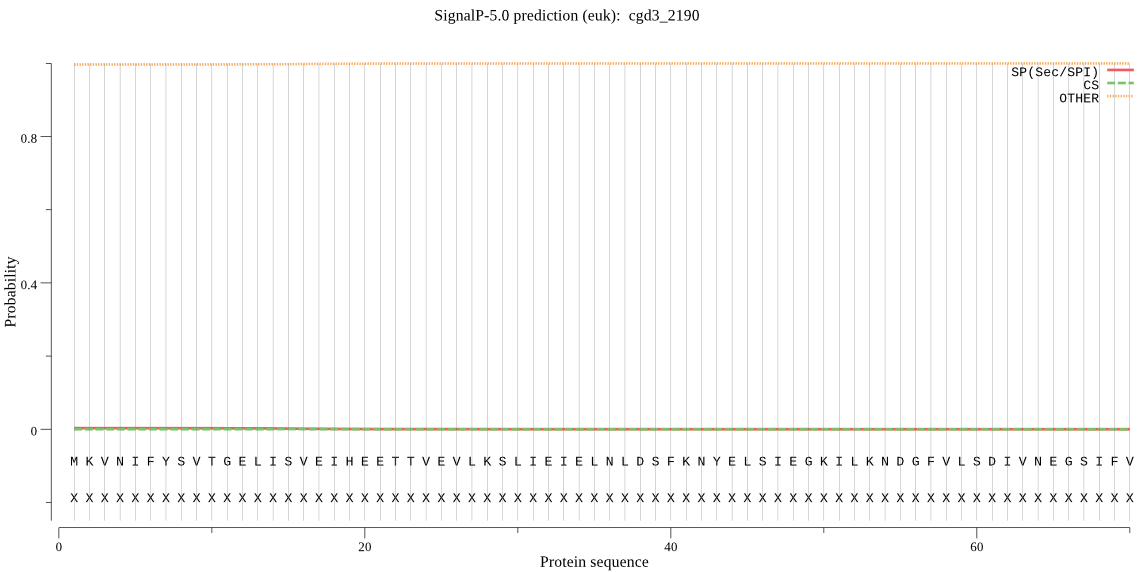
| cgd3_2190 | OTHER | 0.999654 | 0.000304 | 0.000042 |
MKVNIFYSVTGELISVEIHEETTVEVLKSLIEIELNLDSFKNYELSIEGKILKNDGFVLS DIVNEGSIFVLSEKTNKNTFENSNFSLNDRENLIQLLSKELFTKIKEDQSLKSVYYSKSS KYRDSIDKDDIKSFTELIKNDYFSGNLNSISSSSNSSSNLYNLDPLSPEYQRLIEEQVRK QNVEENLILAQDHLPESFTQVHMLYINAEVNGISIKAFVDSGAQTTIMSKKCAEKCNLVR LIDYRFSGIAQGVGTSKIVGKIHVAQMKIGNSFFPFSITVLEESHVDFLFGLDLLKRYQC CIDLHQNALIIGDEKVQFLSESEINSEISQINSNNEYDQCFEQGKKLQLLSLGFSESQVI NALRATNGNTELAASLLFSNETFE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| cgd3_2190 | 167 S | LDPLSPEYQ | 0.992 | unsp |