

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd3_2680 | SP | 0.126084 | 0.870003 | 0.003913 | CS pos: 25-26. AVG-NF. Pr: 0.2303 |
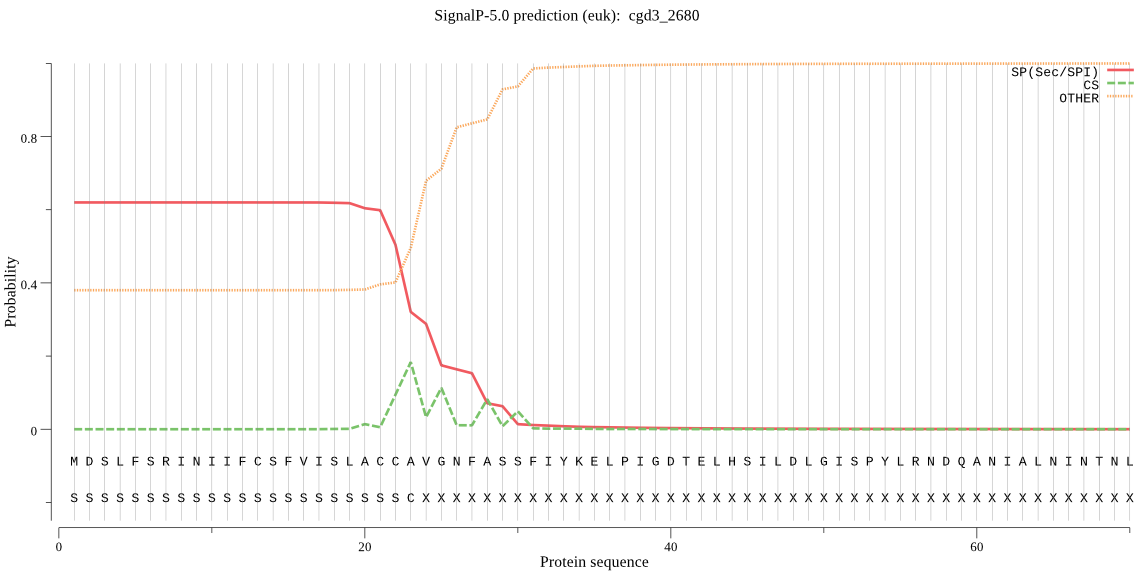
MDSLFSRINIIFCSFVISLACCAVGNFASSFIYKELPIGDTELHSILDLGISPYLRNDQA NIALNINTNLSNSLNWNTNQIFTFIYVSYKNKHQNNYVTVWDDIFSKKKNKTSFSMKGVI NKYPIRDIGRNLRSKSINLNIAFCYMPIVGSIKYHHLKVSTEKKLPVNYFQYK