

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
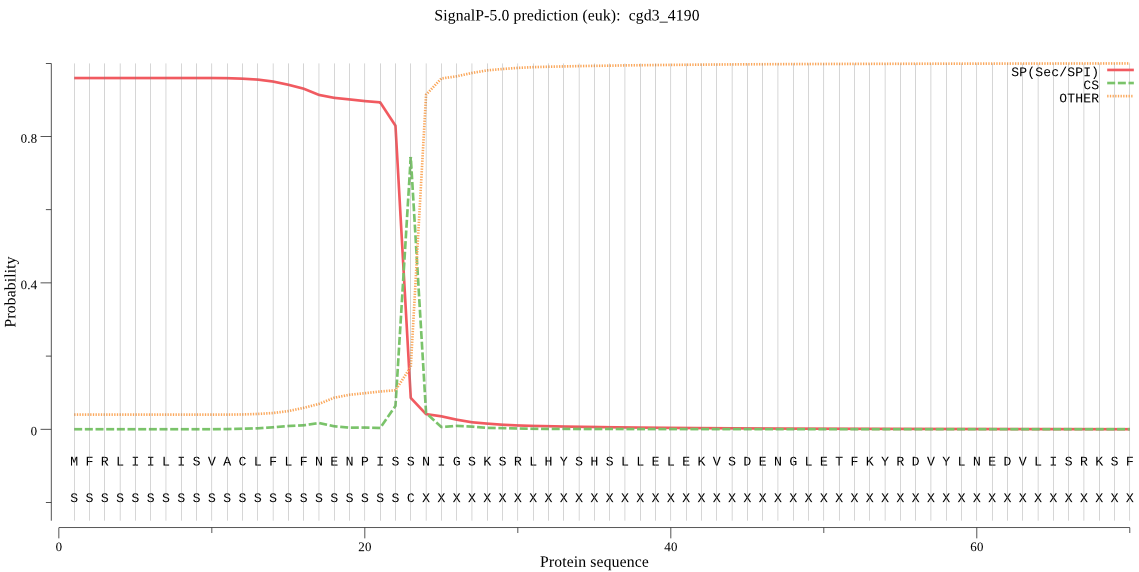
| cgd3_4190 | SP | 0.053952 | 0.944494 | 0.001554 | CS pos: 23-24. ISS-NI. Pr: 0.8104 |
MFRLIILISVACLFLFNENPISSNIGSKSRLHYSHSLLELEKVSDENGLETFKYRDVYLN EDVLISRKSFNNGFTLFNIQQPFESSYSNFRITFNSGSSSDPIQLPGIRYFALFIFSKQV NVCMSKVFKEETHAHFKIKIDATRSTLIFTFLTEKTNKFFDCFLNTIKNQSYLENSSYLI EILQASEEFERILDDPSSIEVDKLMIRMIYGSLPSLGCEQLQISSIFSQSCFKSSIKLDV ELPKYSRQIVNSIFKPHAMSAIFIGKLNNSSLIKGLSSLTKGKLRKKEDLDYNKINIANM AIKNLTSNIVLRNGFHTNILRLYIPFETYDLESLASNGQAFILSILNSRHRSGILNFIFM NRFANNVNSYYYSEYTTTILYLEVNLTKKGLQNIPIIIESIASYLNLIKRTVASDRLFAE AQNLFNYHLTKSTVILSEAIENYLSSHFIMSRPSSDILNSNINTQFIRANVEHFISKMNL DSITVTLHILDQLNPLSVITKKNEKFEIFPVETVEPHTNIRVFIIKTADILSNKLSSLSP TYAVDTFGLQLPNNDLISNISLPFFHSMPIFEKLLNLHKSLDIHYKYLKESGSETIISRT MAYKSITTSGIWFSNNGSFSKSVNLELKFGLRKWSPLLEKFNGEVSSFLILATIHILTAI LNIKLSETLYYTNRLKSNIGFIPAQSFSDIISDPFEIFLVVNAPIDYFTLIMNDVLNTIL SFDSYLTEEIIISAKRAAKSTLLRLYKGYTQFEKDLKIVTQIVSSQHCSFLRLGAFLEND FDIELVRIVYSSLFASPSIYGLIQGNLTPFHANHILNVFIQGLGHEFFSMEINEENSLNF FNKEVYNNDKNDSNNYVPIEINTSILDISTIPNEYKSMYTHKLDFTADASYSTVVLLIGQ LTVPSIIKASAIKEALSNEISSRVSSDKKIDFASSLSVVANNFVVVSLSIRSIEFLAGTL NRILDKELNQSISKVLKSHKSKSYVNKIFENMNFADNVFETKFLLSKISILNLVGGTREY LNKMKPSDISNLLSDIETVPRIMIATQKVNNLKASKTSLDFIPSGYKDIKNDYKCLLEHP NTVFN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd3_4190 | 926 S | SRVSSDKKI | 0.997 | unsp | cgd3_4190 | 926 S | SRVSSDKKI | 0.997 | unsp | cgd3_4190 | 926 S | SRVSSDKKI | 0.997 | unsp | cgd3_4190 | 978 S | KVLKSHKSK | 0.992 | unsp | cgd3_4190 | 591 S | YLKESGSET | 0.996 | unsp | cgd3_4190 | 925 S | SSRVSSDKK | 0.992 | unsp |