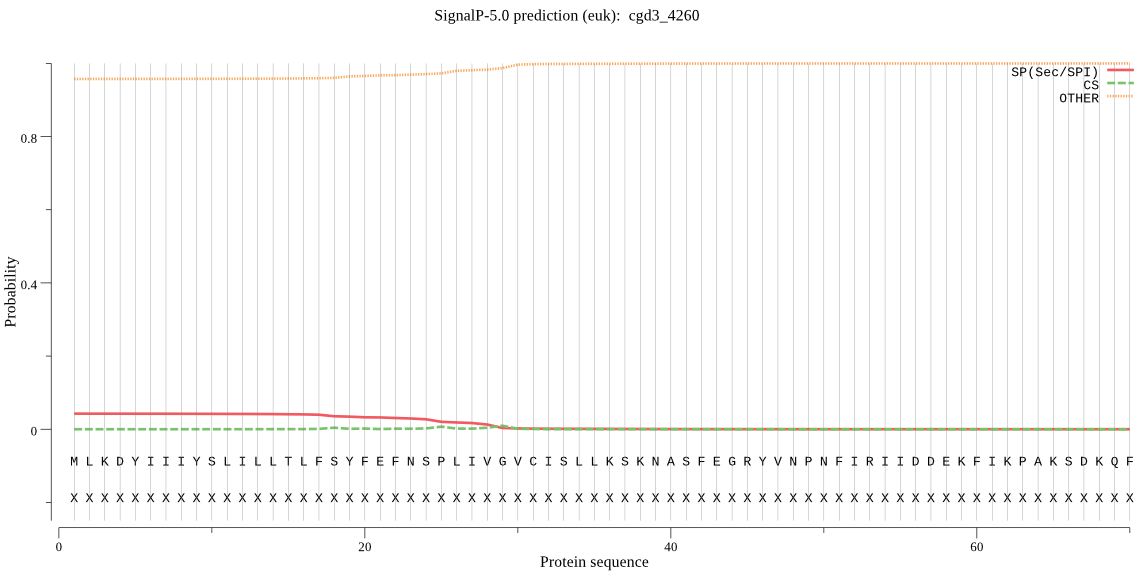
Fasta :-
MLKDYIIIYSLILLTLFSYFEFNSPLIVGVCISLLKSKNASFEGRYVNPNFIRIIDDEKF
IKPAKSDKQFRYIKLKNELEVFLVSHNDTKVSSANIAVKVGSYMEPDSFPGLAHYLEHLL
FINTEKYPELDGFNKLISLHNGYTNAYTEDTSTSYLFSIDSSSFEAALSMFSEFFKSPLF
DENYVEKELMSIENEFNFRKDSLFFRFNHVTHELSDKRSLFGRFSYGNIETLKTIPESQG
INLRDEVIKFYQKEYSSNRMVLALASNHTLDELTQFAYKYFSNIENKNLPVNSIKTPIQN
GNLNPFNTMINQLVVIETLDDSRILKLIFPMKEYMVQHKNKVRTMYIDKLISFDRPGSLG
HHLKSKKLILNMDFSIIDDNLGFTNAVIGFELTIDGEKNIGYILLSFFSVIKFASNNEFS
KEIYDEWKNLIDISFKYEDPTSTSDQSEEIVTYYIKHECKPEDVLYSDYYMDEFDPNIYK
EINSQLTPENLIITLERPDIEDLINMNQNNFNKSCNSISFTEFETVNYNHSKYGIEDLIY
SNIVLDDIRIEKFSKAKFFTNTLSSCLLLLLSNITQNFSIEKYGQSLPTLNPYIPNDFSD
NLNNVSDEKVPLRLSEVIKKYQVQYKNSSFKINNHEMGKYNNFFYYPNKSIVSPKSIIQF
RFIFPLDKITDNFSEFSTDTVTFNLLFMLFLHIFGLVLTRNFFEILSATYSIEQDDVRFI
NDSSRTNQLILTTFGFSDHIEEITSELKSFIQNFSTYITNNDFELQKEKFLIDFKGHILN
PTIYSELIEMNNKLFFNQDYSLETRLSRLETINFEEFIKFSEFYLNNYKLEGFILGNLNP
LQSINIVNAISTTSSLENGNSSVISNYNLLSILSTGVNKSVKLLRLLYSSITGIFISMDH
EDQDYLNVQEHLRPSPPLPMSNSIRDFQKLDPLSLKKGTKIYSYYLSKSKISVNSAIFLR
VSIGYDYMKNYILTNILISIISEEFFTEIRTKKQLGYVVSAKYDIIVDHIYGINFNVVSS
NKNIETLANNVLEFWDDWFYPDSNKITETLFNVAKESFIETLKEKIVNIKGLFSYFSREI
TTKTYDFDWKKNYIEYAEKLTYQYFLDWFKQIYDNSVLFMFAIQSPISEEKSVLDTLSNF
VPQNFTKLTPSDSLFSIENIRTYNQWNVFNAI