

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
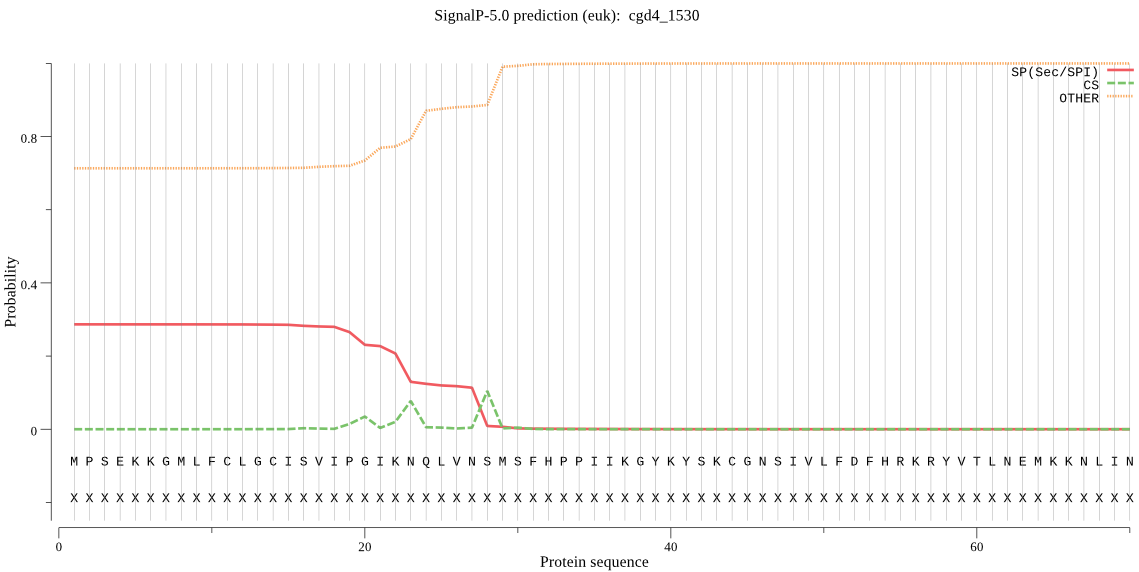
| cgd4_1530 | SP | 0.362587 | 0.632444 | 0.004969 | CS pos: 23-24. IKN-QL. Pr: 0.6162 |
MPSEKKGMLFCLGCISVIPGIKNQLVNSMSFHPPIIKGYKYSKCGNSIVLFDFHRKRYVT LNEMKKNLINIDPGRCQIQVKFSSINSIDFFYYKNPAAKFTIIYSHSNATDIGYLFGHLL DFSHKACVNIISYEYNGYGQSKKKTSEESLYENIKTIVHYSINHLKLPSSSLILYGQSIG SAPTIHFASTYNSINIAGIIIHSGIKSAVSVICNNTNSKSLPWYDAFKNLEKIQKVKCPV FVIHGTADTVIPFNHGEMLYKLSPNKYTPWYVNGANHCNIELNWRDELISKVRQFILYLS PKPKIQSSKSSYMTHDHTLSISRISTLSSQGFENLYHELDDDDDNDDDDDNIYDNYQEYE STSHEYNTNNNKDSCYYREKSYPIPKNHTNNHLNYHNTTKSSNKYFNNQSSRMKIEENNI ISNSSISSKSKFTHSNTYHPNTSNNSHSHNYYSKTLNSAYQTPSSGNNCQISDQVVYGVS IYDDNINLHANMYSGNCRKPKNFTRIFGL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd4_1530 | 151 Y | EESLYENIK | 0.991 | unsp | cgd4_1530 | 151 Y | EESLYENIK | 0.991 | unsp | cgd4_1530 | 151 Y | EESLYENIK | 0.991 | unsp | cgd4_1530 | 310 S | QSSKSSYMT | 0.994 | unsp | cgd4_1530 | 427 S | NSSISSKSK | 0.993 | unsp | cgd4_1530 | 146 S | KKKTSEESL | 0.996 | unsp | cgd4_1530 | 149 S | TSEESLYEN | 0.993 | unsp |