

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
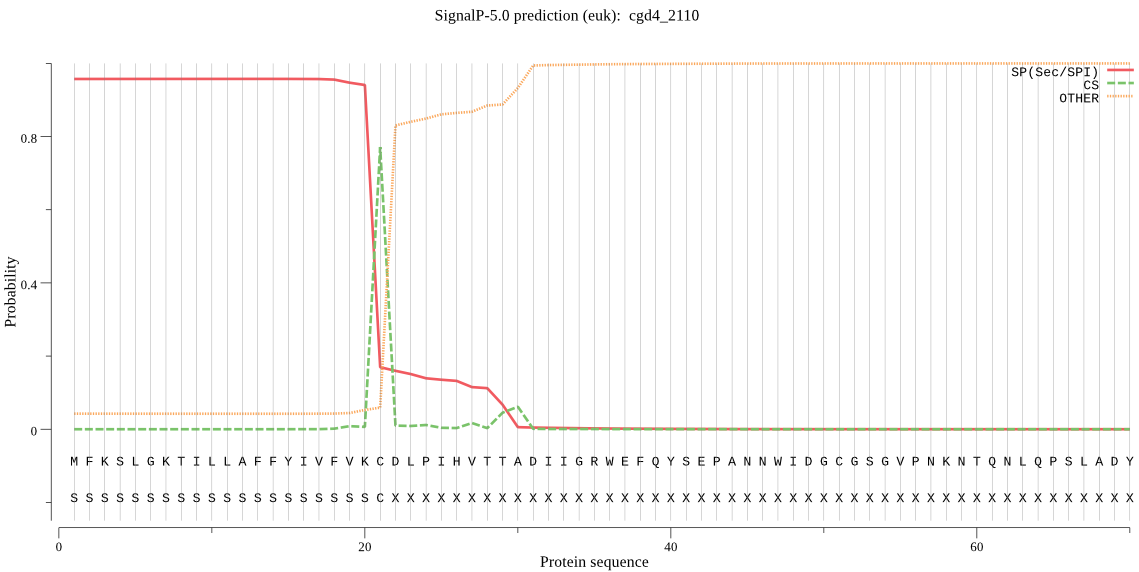
| cgd4_2110 | SP | 0.004445 | 0.995523 | 0.000031 | CS pos: 21-22. VKC-DL. Pr: 0.9546 |
MFKSLGKTILLAFFYIVFVKCDLPIHVTTADIIGRWEFQYSEPANNWIDGCGSGVPNKNT QNLQPSLADYNKWLTTKTNGKLEKLDLILTDLLFSENDYSDLDRSMFPGRSDWTFLAVKD PENGKVVGRWTMVYDEGFEIVVNNMTIFGIMKYNLLNNGNCSAKDGDNETTKGETLCYET DSSQIQIGWYYLRDSKKRGCVSGRKDESYENSLDIIKNRVEIPIKKSFESSIRSLSNSLQ EFAHRHNRKDSKNWRARSNFSLTDVAFSHHSYYMNAVNFRKIHPFRIMGSQHSSFIEMFK DGVGSVDNYDNIVDSIFACNVVRRKTDEVKLPSSFSIGDPFTDEKYSDFPFNQGNCGSCY AVSSVYILAKRAELKLNKLTNGASREEKILLSPQSVLSCSPFNQGCEGGYPFLVGRQAEE IGISSEKCMGYYADSNQECNFSPFITPEIEDRIYCEEGERMYAEEYGYVGGCYGCCDEDR MKEEIFKNGPIAVAMHIDTSLLVYDNGVYDSIPNDHTKYCDLPNKQLNGWEYTNHAIAIV GWGEENGIPYWIIRNSWGANWGKKGYAKIRRGKNIGGIENQAVFIDPDFTRGMGLSLLNK YQNSTAYRTNLPDIKDSPQGSEKVNVEIVNGGVDQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd4_2110 | 195 S | YLRDSKKRG | 0.998 | unsp | cgd4_2110 | 195 S | YLRDSKKRG | 0.998 | unsp | cgd4_2110 | 195 S | YLRDSKKRG | 0.998 | unsp | cgd4_2110 | 208 S | RKDESYENS | 0.996 | unsp | cgd4_2110 | 99 Y | SENDYSDLD | 0.991 | unsp | cgd4_2110 | 162 S | NGNCSAKDG | 0.995 | unsp |