

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
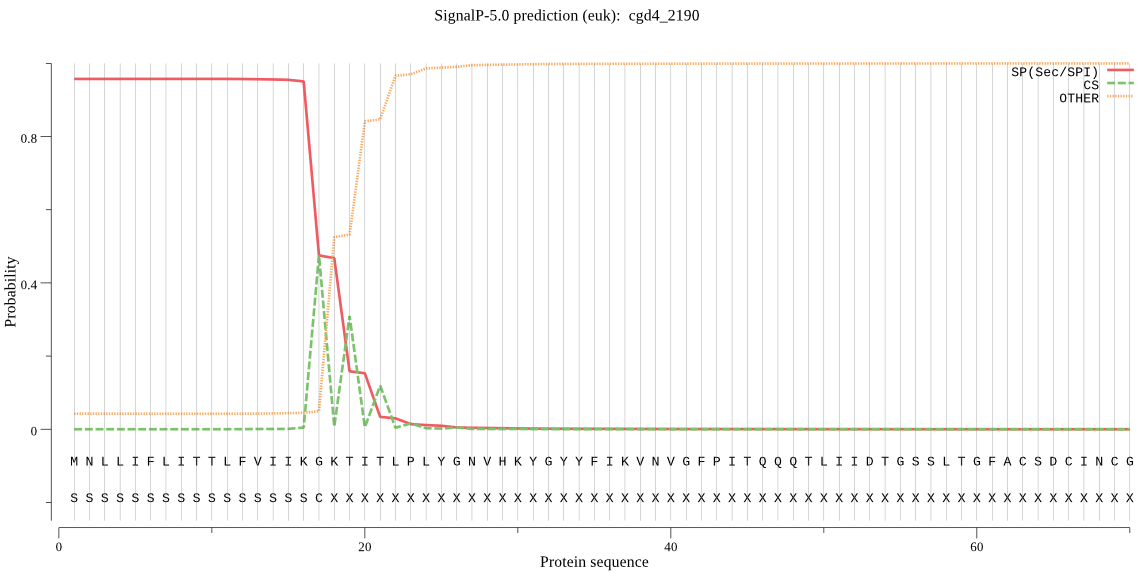
| cgd4_2190 | SP | 0.003477 | 0.996505 | 0.000018 | CS pos: 17-18. IKG-KT. Pr: 0.7361 |
MNLLIFLITTLFVIIKGKTITLPLYGNVHKYGYYFIKVNVGFPITQQQTLIIDTGSSLTG FACSDCINCGTHENKPFNINLSDTSNIIKCKRNNTPNNETDIINKSIHGRISMNYPNYNK SFLNNKCVYDIKYSEGSRILGYFFEDFVEFENKLSSNLEIRQKFKNKFVFGCNIIENNFF KFQKASGIMGLANFSNKEMNQIINYIFKSGEVRKTDSDKIISIFFEKDGGKLTFGSTCFD QTKMMNYPFENYNITRCINDERYCAYISKIEVDSNTRELDTKLNERLFKAIFDTGTTISI FPARLFKKITRGLFNNVSKYYPKISGHDEKDGLTCWRMLNGISTDKFPNIKVVFNNNRNK LTEQLVINWPPESYLYLNKILEGNIKVYCLGIASNNLINSEIGADKNGENSSSNEIILGA TFFIYKEITFILNENKIMIRYNYLNSKDRNGILPKTTNNFRHKLNNSNSDIGATNYYEGK LTNAHVRDIHKNVLSKILVKRYVVLKNKKYEIWGILISSSLIIAILFSFLTIFHKFFRKF NFSVKSNYGI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| cgd4_2190 | 325 S | YPKISGHDE | 0.992 | unsp | cgd4_2190 | 543 S | KFNFSVKSN | 0.992 | unsp |