

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd4_4160 | SP | 0.216077 | 0.773210 | 0.010712 | CS pos: 23-24. LKS-IS. Pr: 0.2629 |
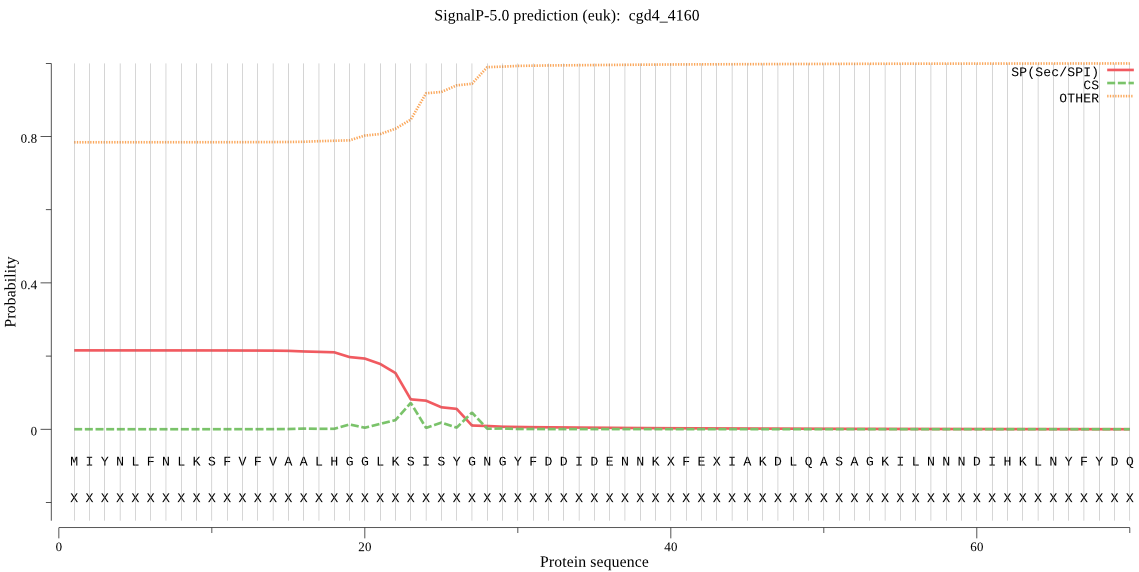
MIYNLFNLKSFVFVAALHGGLKSISYGNGYFDDIDENNKXFEXIAKDLQASAGKILNNND IHKLNYFYDQIGNIAKTVYPIKGGFEDWSLYGYNVSHILCSDYIQKSHSANKGGSLTFLI ETDHQKTPNQDTLGSKSDLFSLIDLDKLSIQPSHITRNIRMLLKLTEYTYPDIIFFNKPP STVYFGQTFILYIAIIGCYSFSNLRIKLENNNNNNXNNNNXINHYXNTXLNSEQQPIYLN FYVNEGLDGLQNTIYYKRCSSLFLNNQEIQDILDNXSXYTXXDKCNESNLLTXIVNRSFR VKNKCKSIYQFNKFXPIXILXRYRMRQQFLIL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| cgd4_4160 | 141 S | SDLFSLIDL | 0.994 | unsp |