

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd4_620 | OTHER | 0.748431 | 0.153499 | 0.098070 |
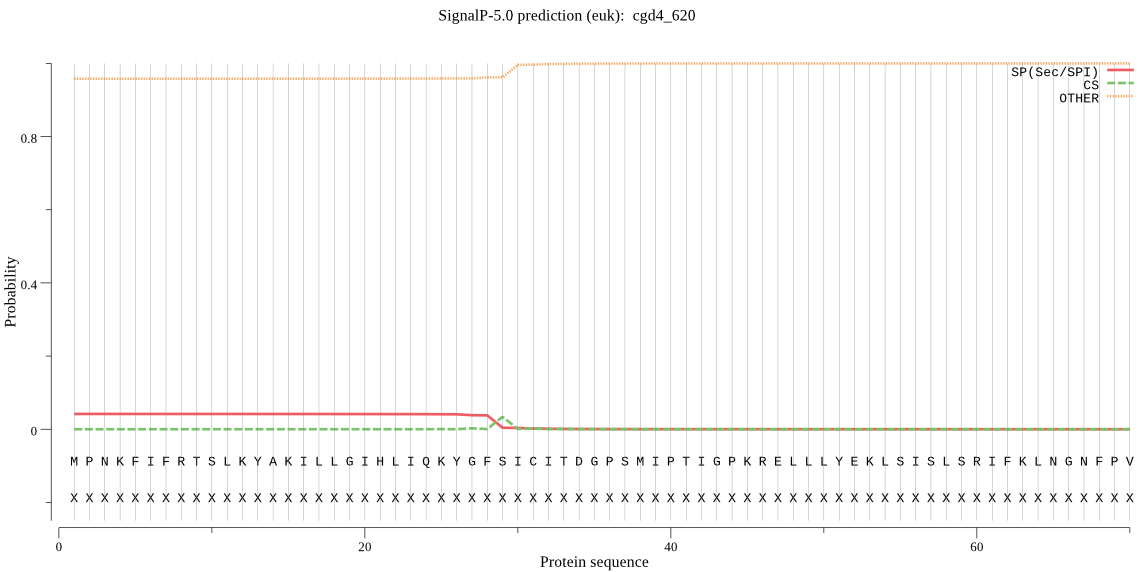
MPNKFIFRTSLKYAKILLGIHLIQKYGFSICITDGPSMIPTIGPKRELLLYEKLSISLSR IFKLNGNFPVNRNDIIIANSVENPEILVCKRVIGKEGDIVTVFPNYTLVSKFNNCNFIDF IHKRHSCFQMKIPPNYFWIQGDNFNNSRDSRNYGPIHESLIIGRVIYKLWPNPIFSKLSA IIRRKMDQINTLEN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd4_620 | 126 S | HKRHSCFQM | 0.99 | unsp | cgd4_620 | 126 S | HKRHSCFQM | 0.99 | unsp | cgd4_620 | 126 S | HKRHSCFQM | 0.99 | unsp | cgd4_620 | 10 S | IFRTSLKYA | 0.994 | unsp | cgd4_620 | 80 S | IIANSVENP | 0.991 | unsp |