

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
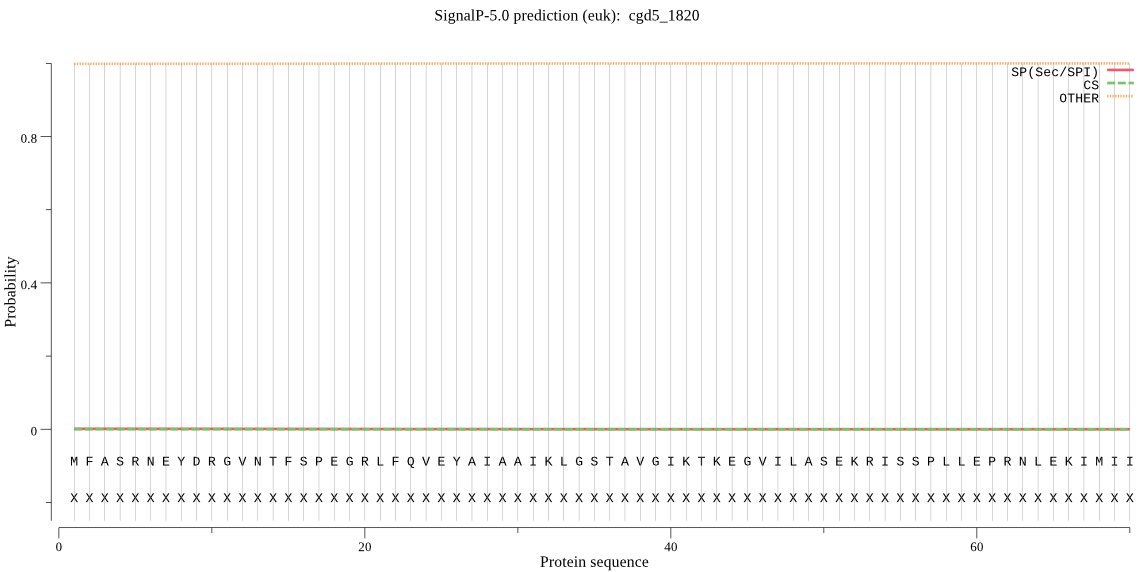
| cgd5_1820 | OTHER | 0.999631 | 0.000110 | 0.000258 |
MFASRNEYDRGVNTFSPEGRLFQVEYAIAAIKLGSTAVGIKTKEGVILASEKRISSPLLE PRNLEKIMIIDRHVGCCMSGLVADAKTMIDHARVESQNYFFTYNENIPTQSVVQSISDLA LDFSDIKEKGKKKSMSRPFGVAMLIAGADSDGSSSLWMTDPSGTYTQYSAAAIGTAQEGA EAILLENYNSNMSLKEAEDLALIVLRQVMEEKINSVNVEVAAVKEKKFIIYDDKEIQRVL DRLPPPTHIVPSQLS
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| cgd5_1820 | S | 255 | 0.565 | 0.029 | cgd5_1820 | S | 252 | 0.509 | 0.237 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| cgd5_1820 | S | 255 | 0.565 | 0.029 | cgd5_1820 | S | 252 | 0.509 | 0.237 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| cgd5_1820 | 134 S | GKKKSMSRP | 0.992 | unsp | cgd5_1820 | 193 S | NSNMSLKEA | 0.997 | unsp |