

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd5_3220 | OTHER | 0.959425 | 0.000302 | 0.040273 |
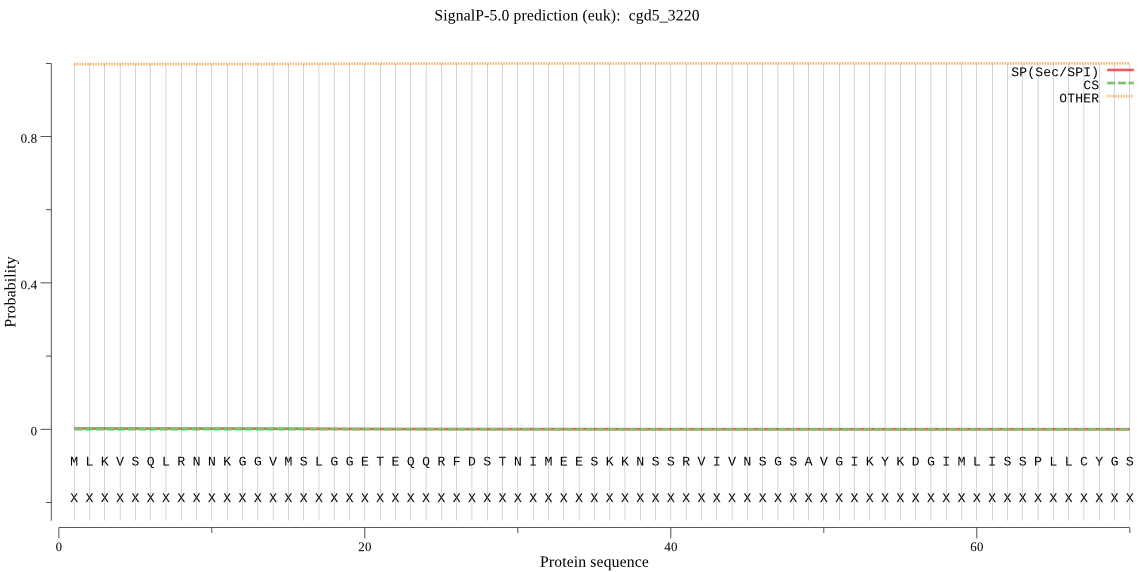
MLKVSQLRNNKGGVMSLGGETEQQRFDSTNIMEESKKNSSRVIVNSGSAVGIKYKDGIML ISSPLLCYGSMKMNSDISRFHVIGSGMIDLYDERQKLKSKDEDMSSVNTDKRPLVRKLTE VDEDAKVRFYEGKLSVIVSTGEFSDFQNIIERVEEKAAEDFFEGNFKSAKEYSGYISAVH YQRRNKMDPLLNDIIVAGLRKDGSKEIYSVDPLGTRFDEDFIASGMAEYLAITLLRDKYK PDMSFDEAKLILEDCMKNSFYIECKGARMVQIATINQDGTRIYPKYPLDVTWRHDLFVKS NLDRKDYVGF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| cgd5_3220 | 99 S | QKLKSKDED | 0.996 | unsp | cgd5_3220 | 105 S | DEDMSSVNT | 0.991 | unsp |