

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
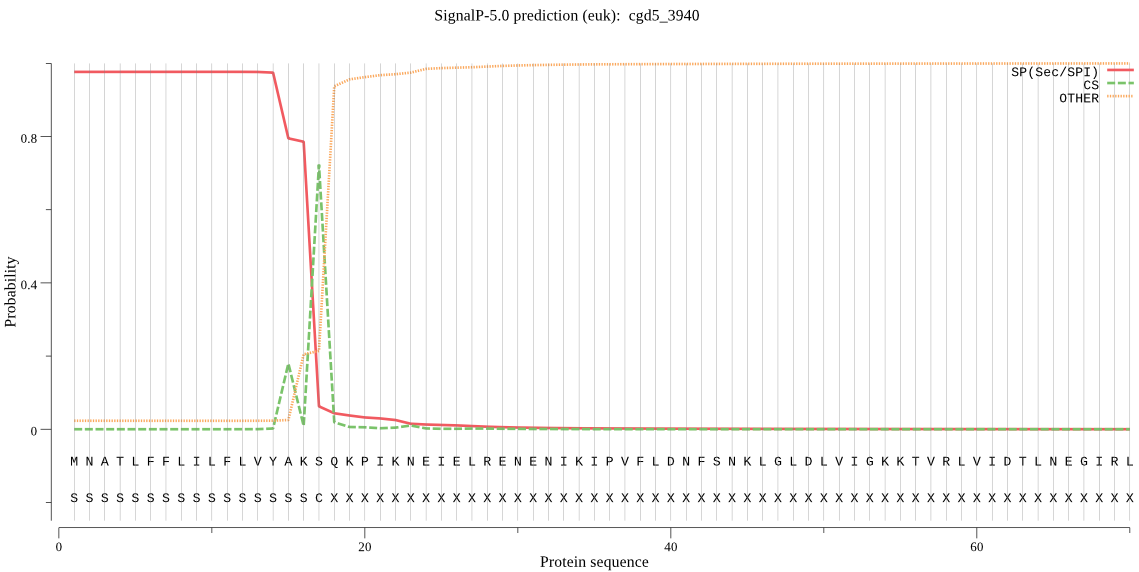
| cgd5_3940 | SP | 0.000929 | 0.999066 | 0.000004 | CS pos: 17-18. AKS-QK. Pr: 0.8183 |
MNATLFFLILFLVYAKSQKPIKNEIELRENENIKIPVFLDNFSNKLGLDLVIGKKTVRLV IDTLNEGIRLFQDGTEACNKGKLSYSFRSGTGFEDDSEISRIGKSQSNNDSCYDPLSSDS ASWCYNKDHCRISYYMDSYTCEKKKEFPDTRENGIFNAFFHSNERVYDGISKFEVMLEGN ELVIIPWMVPELVLQEFPIKLIKSDLSTGNGENWPSFHNFDGFIGLVGSSMSCRGISIWN DILMLYNASRVYFDVNFENPNSSFLYLNNLPEYWDPISIYWSEPKQSGFVYDDALSLFQI YNFEICGSNIMENISSNWLAALDLSSTCLSLPPFLFARLMSWLPLDCPEYSINNDKLELD KSKENYSQNNELLPYSSLCIVKKKPLPVISFWLSQTDSNDGKVEPIKIFLDDYIIHFNNI DYLCVINSTLGTFGKINYIPYSEGNSYNYFTPYYSSSNTPLILLGSLFLKSYGIVIDNNS GKVGFYPKYKYKGYNSNTIEHSVNSNCAPKKECLLENGFKYDSSINECIPPNCSDWLLYE FNTETNTCELNSIFPLVLAFIIVLFTFFEIHIMYLKQYILDRSSEISNS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| cgd5_3940 | 207 S | KSDLSTGNG | 0.996 | unsp | cgd5_3940 | 584 S | LDRSSEISN | 0.994 | unsp |