

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
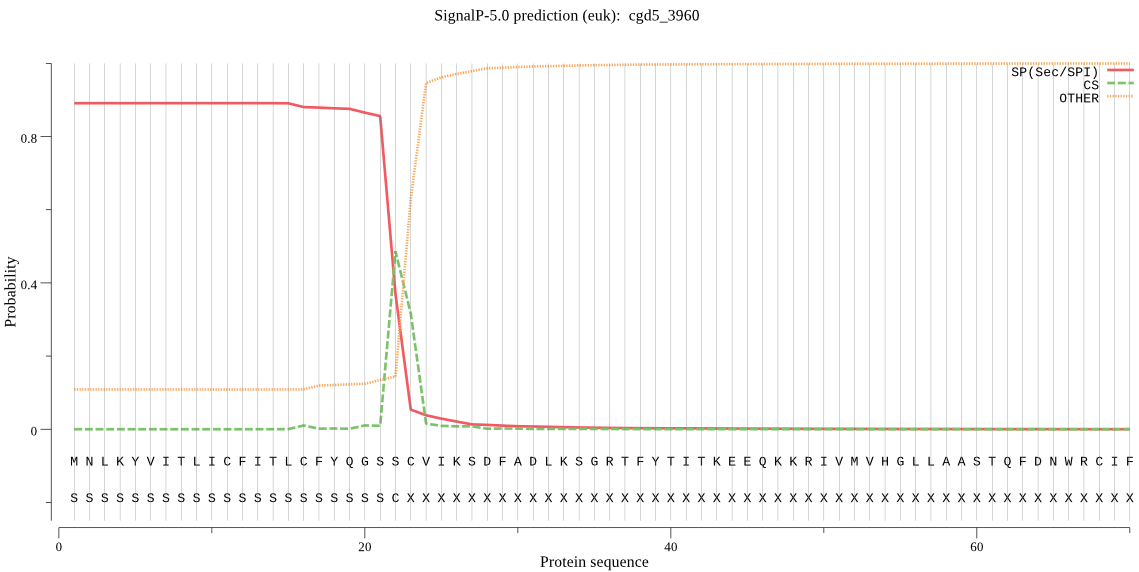
| cgd5_3960 | SP | 0.015867 | 0.983940 | 0.000193 | CS pos: 22-23. GSS-CV. Pr: 0.6892 |
MNLKYVITLICFITLCFYQGSSCVIKSDFADLKSGRTFYTITKEEQKKRIVMVHGLLAAS TQFDNWRCIFSHTGYQVLTYDLLGHANTEWKLPGFFSQKRFVDQLNELLKHVGWVDSDNK AVEKISLLGVSMGGLIIINYALEHPDHISNLIPMCPPGIMTKYDFPKLYRLSNSSLVSAI KNIHNSKRVFRCGFYCASKLGYVKLGKQTKEEKKATTNVFHQMFSTYIKSGGDGNLFDRH LDFERLSKYENTFKIIFFWGMKDDVVPFPPALEFLVKHFSSTPIVVYPNIKHIPAYLMYS PALVTLDFLESNFTVGVPLGQVKELKYFYDNEVNIVTGLNFTISNEIIVKSNKENFTESY LDFMDY
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| cgd5_3960 | 247 S | FERLSKYEN | 0.997 | unsp |