

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
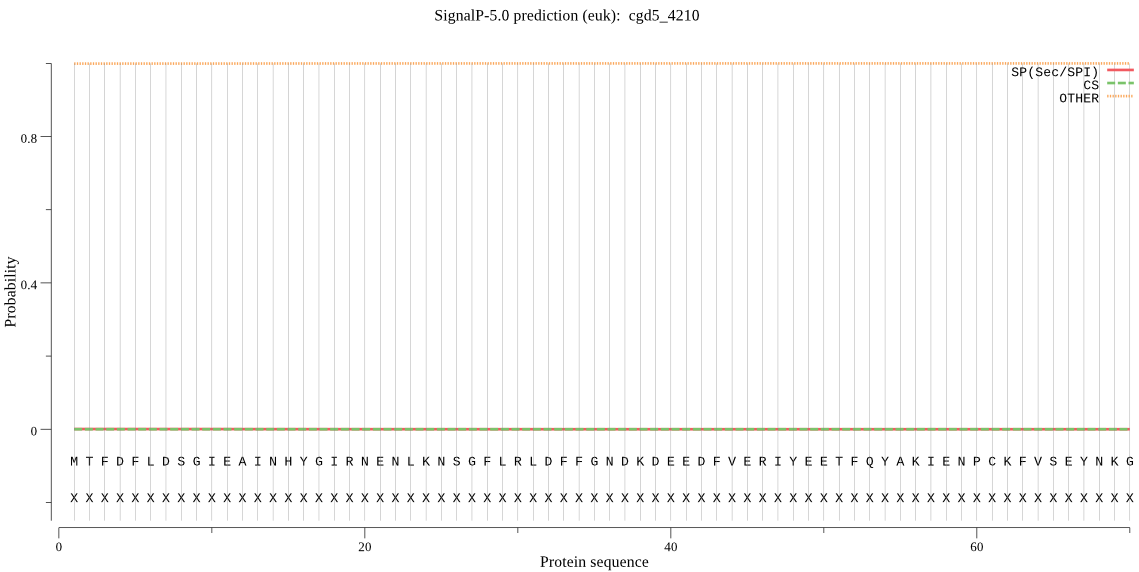
| cgd5_4210 | OTHER | 0.999904 | 0.000081 | 0.000014 |
MTFDFLDSGIEAINHYGIRNENLKNSGFLRLDFFGNDKDEEDFVERIYEETFQYAKIENP CKFVSEYNKGEIRKMMNRGTTTLGFIYQGGVILAVDSRASQGSYIASQEVKKIIQINDFL LGTMAGGAADCSYWERVLSKLCRLYELRNGERISVAGASKMITNIFFHYRAYDLSAGIMI AGFDKDGPHLYYVDNEGSRVKDCKFSVGSGSLYAYGVLDSGYRYDLTDEEAIDLGKRAIV IATNRDGGSGGLVRVYQINKNGVIKHVPGEDVSELHYAYSKKQGTDPTKM
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| cgd5_4210 | T | 288 | 0.513 | 0.276 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| cgd5_4210 | T | 288 | 0.513 | 0.276 |