

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
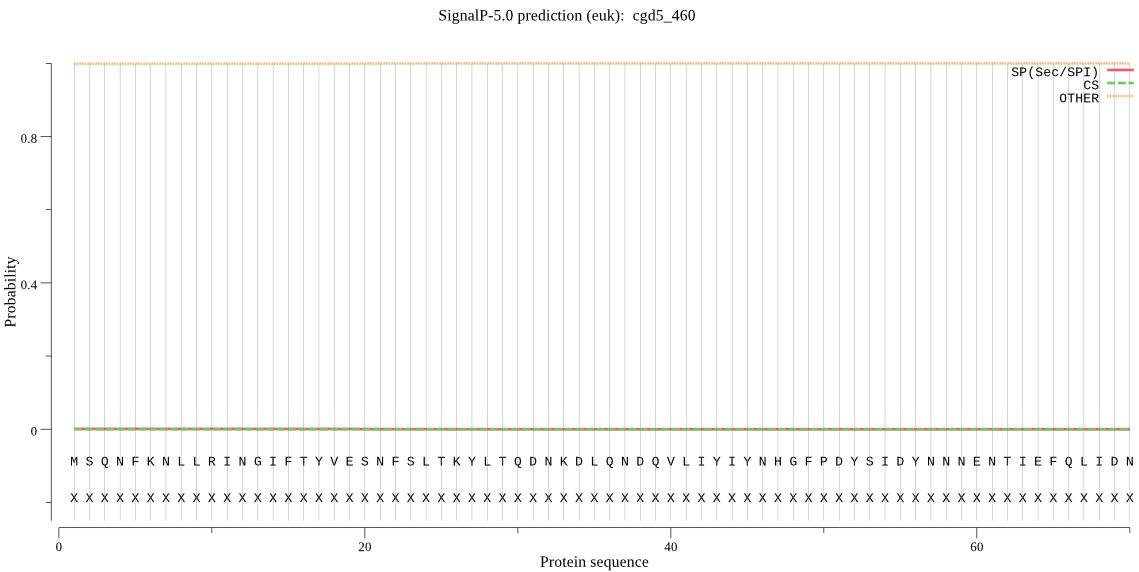
| cgd5_460 | OTHER | 0.995357 | 0.002221 | 0.002421 |
MSQNFKNLLRINGIFTYVESNFSLTKYLTQDNKDLQNDQVLIYIYNHGFPDYSIDYNNNE NTIEFQLIDNYLCHSEAYKLEIIKDLIDENIKLSRFCGKMRDICLSENHINKLKEKSQID KFLFVSFNTNGIGKLSQRGEFYDKTLTKDMYDIETIVECFSNILFKNSKLIMIGISTGAF LTFSYSTNYTRKDIKCENEYFISNNNLIGIICIACVDNIPESYILDFNNEQLNEFNELGY TKINTKIPIVKINHINKIKDDLKLISRDYLESYNIFPTYNFLLENKEKVVNYPILLIHGT DDQSVPFQMSLNLLNLNNNFSNKNNRYLRLLKINNGNHLLTNSKHIKKAQNEISNFVFEI L
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| cgd5_460 | 117 S | LKEKSQIDK | 0.996 | unsp |