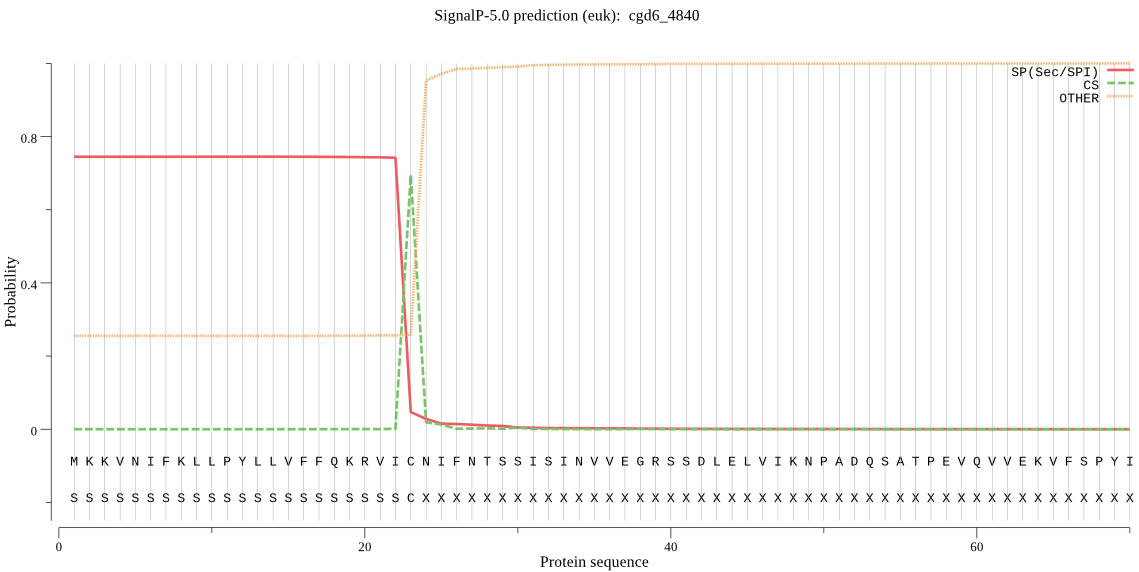
Fasta :-
MKKVNIFKLLPYLLVFFQKRVICNIFNTSSISINVVEGRSSDLELVIKNPADQSATPEVQ
VVEKVFSPYIMKQQKPPEEDYEQEHILGEFSEMGNMDDEDDYYYQEKDSNQEFNSEKVSR
GQGRRSKVIPGRLIIKYKEKVLSKAVKELKVAGIMSKNGQYPKLDGVAFLTRVVSALSES
ADELVGIKSATVLEALDMMIVEVFNNSKASEYISKLSASFLHSDEVEYVEPDQEVRLADF
GISSDSGAIYPNDPKFSEQWGMYNRVANTDSKVLMAWKHLGINTSVSKGSGVYYTPENHP
KREVIVAVIDTGVDYTHPDLVENMWVNEKELYGRPGVDDDMNGYVDDIYGYDFANNRGAP
VDDEGHGTHCAGTIAAKGNNHEGISGINWKGVKIMALKFLRGNGVGFLSDSVKAINYAIK
MGANILSNSWGGGSFSQATYDAIKRSIDKNMLFVVAAGNDRNNNDIRPTYPSAYQLPNVL
SVAAIDIHGRLGVFSNYGHRSVDMAAPGVDILSTSANKGYKRLSGTSMACPFVSGAAALL
YAFDPSMTFDQAKTLILRSVTVLTTLRNSVRTSGTLNIHRAVQMMSQEAYNQENGKEFPR
IGTSWITFKAPDTIGPQSSSILKLKAFGEKPGSFTSTLVVKLVDSNDREVTLAQGRLKTI
INVVSIPKPKAILPNGNSTNLGLSFMGLEGTKTLIPISNSGIGTLEYMVEFVNMEGELGS
KRKGKFKVYPPLGTILTVGEDQTNKDLQVSCIPHSLEGFISGHLMLRYNKGGMSSSSSSF
SSNKISAKSTKIPYVRSNLFTETSSIINTLSNNIISPNSNYILSSSSSSSFPVGTERLAI
KLYCKGYGIEIRPPSINRVIRIPETTISGFILFKGLENSFKPRIYLRFKSLRDPSSTGFE
RESKRKDYSRSNTSIKDLSPSKSYFKMVLSGENLPSKKRIIHRLEGMDSEEGKNKLYVSL
DDVILPKANVKFDWVNIGINQETILTDLSNSDDGVQKVNLPRSWYFLGEKIDEIYVSVNG
FISFSPIYSEAFVPALPSYAPPHGILAPLWTDFTTRGRRENTKIVTFLKTEEDEEDFGGG
FGSGVGSRYRREERLEYEEEDDGEEKEEGKKRRWEFVIEWRNVFLKSEQYNSKSATFQCV
LNSNGSVKFNYLSIPWEDLEDSRGISSSEKGSLAISRISSSTMLGWESIDGIRGVGIPCT
EKFPTSEFSIELYPENPKVPWFQLSETIVPPLQRAQMIYKVGWKAVIPSTFEDQTFKGQI
QVSSSDGAASIIPVSITAIKEKVNQAQSSSKDLEEEKDDDEEKEIGVSREARDVEENEFG
RLDS