

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd6_660 | SP | 0.486307 | 0.513386 | 0.000307 | CS pos: 25-26. TEK-LT. Pr: 0.4930 |
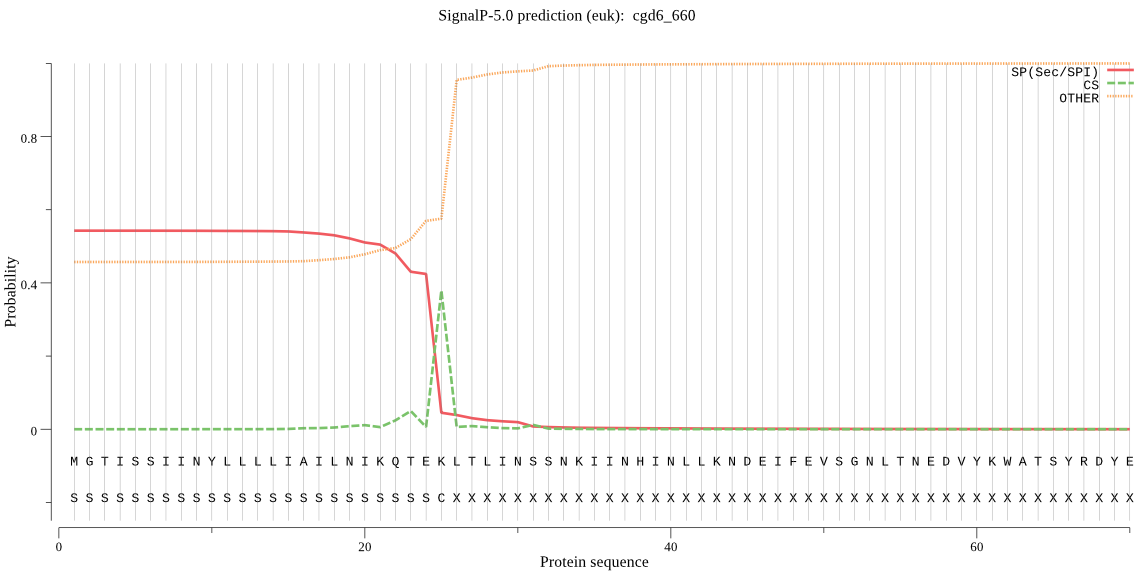
MGTISSIINYLLLLIAILNIKQTEKLTLINSSNKIINHINLLKNDEIFEVSGNLTNEDVY KWATSYRDYEQDSEHDHENDYDADIEDNIDDDDDDYDDDDHHHHRDRIHNNKYEDHNQYQ NSLSLNKSKLIYSDSESFLINPKLKNIYDFSNNKQIYKTINVLNVNGEIIPYDVFITNIT IPLFEVKNSLFVCRIRIGEPEQEFWPIIDTGSSNLWVIGEECNQSSCQKVKRYSKYISKS FKRISKYDNISVIFGTGKIYGKLILETLKFDNFQLNKHVIGIIEQVENTENSKIDIFDAI ELEGVLGLGFTQMSSSKKLQLPLIERLQSQGLIQNNIFSIYINDYRIKPINNNDKLSKPC AILLLGGIDQNLFEEELHILPVIREHYWQVELESLYIGDTKYCCDYGSLAYEWENLENEH LYKKFGHYEDPPMDVYIYKTFNKSIDKTNRTPGYVIFDSGTSFYTLPNFEYNHFIKEYQP FGDCSQIHLDSKKIDPKIIQHFPNITYTFKDGFQLVIPPELYLTPNEQGKCKPGIMMIDV PGEYGHSYLLGSLFMRSYYTVYAKNIPKIGSAVGIAKAKHNKDTRKYIYSKLSSINETFV PPDDDEAELYWTQINGRLSENWKFPPILGNLFF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd6_660 | 96 Y | DDDDYDDDD | 0.991 | unsp | cgd6_660 | 96 Y | DDDDYDDDD | 0.991 | unsp | cgd6_660 | 96 Y | DDDDYDDDD | 0.991 | unsp | cgd6_660 | 234 S | VKRYSKYIS | 0.994 | unsp | cgd6_660 | 240 S | YISKSFKRI | 0.991 | unsp | cgd6_660 | 245 S | FKRISKYDN | 0.998 | unsp | cgd6_660 | 292 S | NTENSKIDI | 0.991 | unsp | cgd6_660 | 65 S | KWATSYRDY | 0.997 | unsp | cgd6_660 | 73 S | YEQDSEHDH | 0.998 | unsp |