

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
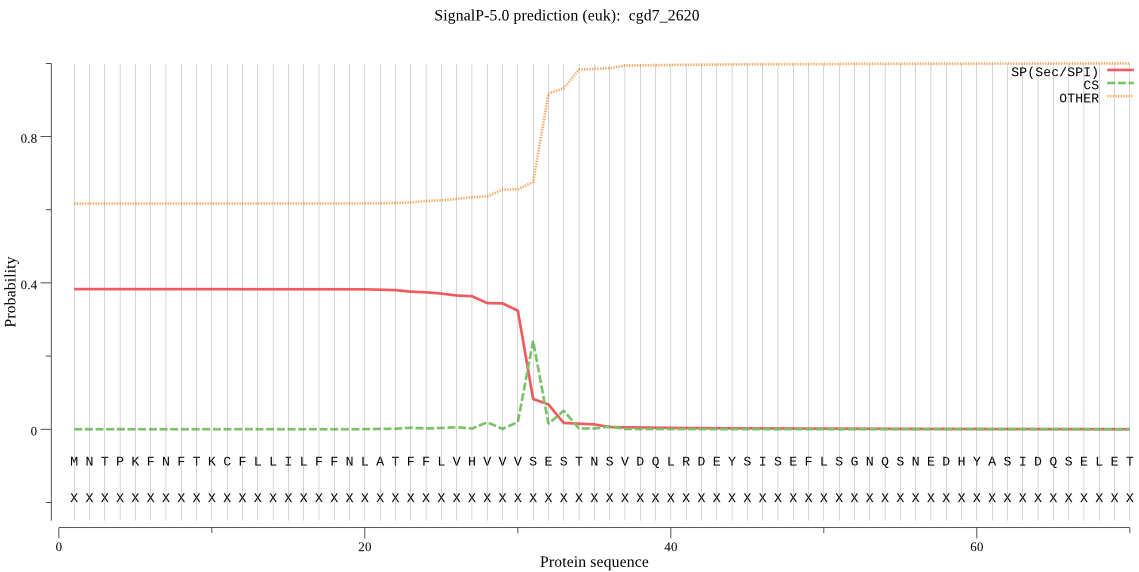
| cgd7_2620 | SP | 0.283495 | 0.716049 | 0.000457 | CS pos: 31-32. VVS-ES. Pr: 0.8210 |
MNTPKFNFTKCFLLILFFNLATFFLVHVVVSESTNSVDQLRDEYSISEFLSGNQSNEDHY ASIDQSELETRSSFDSNRESLNAHLESKLTGVIKQSFVEFQKGKLSKNDVDIEELIFEIV KLALDGNGSRVLPSLELIDFLQKIIETSKYNTVTTLDYILAFISKEKFDTSITHDINRLR NFTEYLSYLLKYNDNGPNRMINSGKETLSLIKTRIHSSIWGMEELIRTNKIMDYEIQNLG GKTTYSSQPVIGEITYEINSNQPRAVFSYKLVKQTIIEMLGTNINKVHSNLKRIYGKITY SGSSKSNKSISLNLLYAIYLSAIMINDLYNQGLSSNLLSPGFIFMVLLRDDIEHLPNNWE ERLIKRSISYGINETDIKIQDSKYSNLESIITKGFTHSECNASVLRSNIQVVFRKYGIDY FKFMNEALKKYKTFFVPSTDGRINSISWFNNYSVNGYTNKLNKKWFVDINELVIENGVTK AVDFMDEFHLLEISLSRSGLSSAIIVSESKLLKRTLVEYLAYRIISGNSSIDLRGYRIIS IHLESLLESCKNTKKSLTEQIKIKFDELMGAYDGKIIVFTDNLFSSFETSTGSKRLYDIM KHYIVRGTLKVIATLSNENYKILAEKEIEVKSIFYTIEMKELNGIVSEVFISGLRYQLEL STGIFINNDVIRVSVLMCHKYIENCVLPDDAVELINFAISMAKNEQFLEIPSAIHGIEDF ISHSRIGAQVSKMRDYESNSIISSNRSRLSEVLAMHLRMRNRIVSLALKIRPYLTNFRYL KTQLYFMMLINLNYQTSCVNKPVEDEFKLLENREKIIPNFSEELSVTQLTSLYRKNLLKY KQINNKSYNESINDEVSAITESQLDPMFFDDIHQVVKDKQKEVADMKSLILEMAFNYNSL YSPLGMGEIDASHIAFIISERYGKSITKLLDEIEYRKLPERMKDRLSEILSKYVIGQKSA IDYVSFHLGVELLRDNKNNPRCLLFVGPSGSGKKTFALALQTALTESSVLFYDFSYDITM YSHFKILKSSDFADADAAKRLLGDNPKTGIISEELKQTGKTVFFFEHIENMHPDVIKLIL DILHNKDNINHKIGNLGSSTFILTTKVGSEIILKNPDQIDRIKIRVDIIKKLRETFSSDL VRLIQNVVLFRPFTKEEALKILELNFSEFSVLIKKNYSTIFIQPSLLVLNKIVDKKYSPE LGYKSVLDFFEKEVKEKVLQLIKKGILVPYTTFKITIRDISSVEDPNGFKIDFAIVKKSI NSR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd7_2620 | 369 S | KRSISYGIN | 0.992 | unsp | cgd7_2620 | 369 S | KRSISYGIN | 0.992 | unsp | cgd7_2620 | 369 S | KRSISYGIN | 0.992 | unsp | cgd7_2620 | 529 S | ISGNSSIDL | 0.995 | unsp | cgd7_2620 | 556 S | NTKKSLTEQ | 0.995 | unsp | cgd7_2620 | 947 S | KDRLSEILS | 0.994 | unsp | cgd7_2620 | 1241 S | IRDISSVED | 0.996 | unsp | cgd7_2620 | 1242 S | RDISSVEDP | 0.996 | unsp | cgd7_2620 | 60 Y | NEDHYASID | 0.991 | unsp | cgd7_2620 | 76 S | SSFDSNRES | 0.995 | unsp |