

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd7_3660 | OTHER | 0.999568 | 0.000110 | 0.000322 |
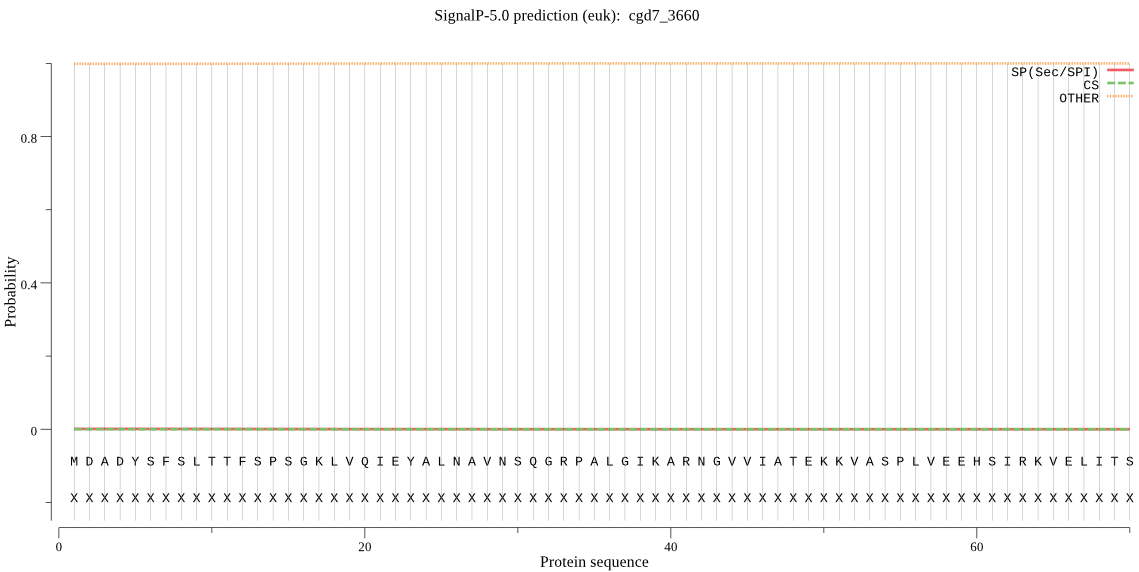
MDADYSFSLTTFSPSGKLVQIEYALNAVNSQGRPALGIKARNGVVIATEKKVASPLVEEH SIRKVELITSEIGCCFAGMPADFRVILKKSRKIAQVYYNTYREQIPVCELVREIATVMQE FTQSGGVRPFGISLLVAGFDSTRGPQLFQVDPSGAYFGWKASAIGKDMQNAKSFLEKRYN PDVEIEDALHTALLTLKECFEGAMNEDNIEVGIIGEDKNFTILSPQEIKDYLGEVE