

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
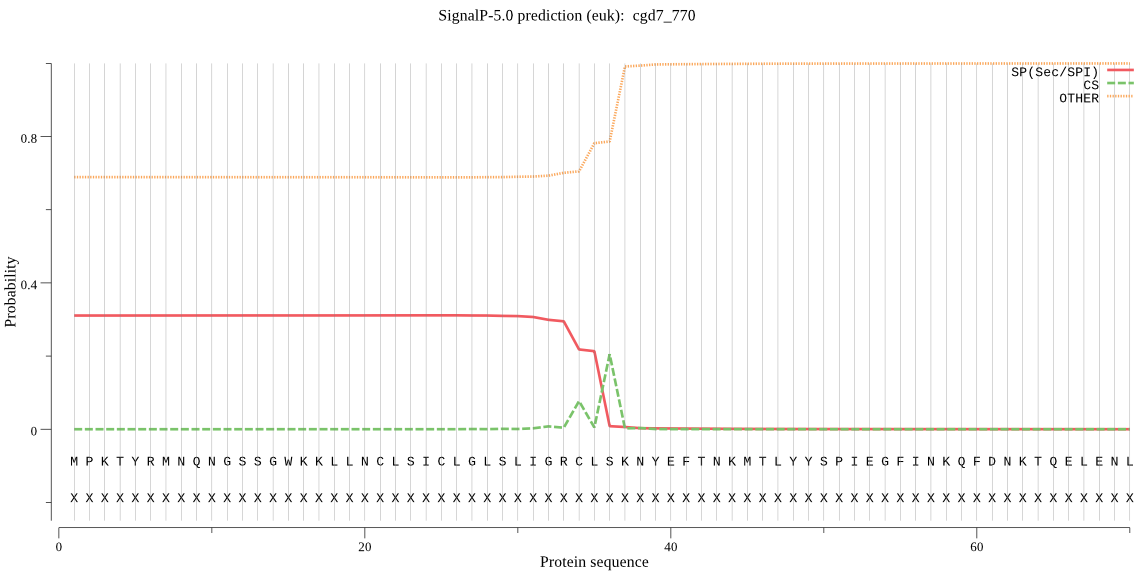
| cgd7_770 | SP | 0.420370 | 0.550828 | 0.028802 | CS pos: 36-37. CLS-KN. Pr: 0.6850 |
MPKTYRMNQNGSSGWKKLLNCLSICLGLSLIGRCLSKNYEFTNKMTLYYSPIEGFINKQF DNKTQELENLKFIIGEPWEGFEIAKVQLNSSSINNDTKTMESLDGKNNVVFGKYKKLEIN YPLGSKKKLIALQNCKTGVTALINYSPMRIKTEFSIYLSFGSNIDPINNQGLTYLMGFIF LNMASEVDNYLFNSGEYIEKQKDNSLTSVTVSSLYTKLTVTVSDDKAIEALKRFINSVGY SGNVDNMTSTSKNDDGEFYIKNRPLTTKNLLNSLCRLYYIYKSCLRDEEYRISFIKTSMR AIDPKKSKEEESSSFGSSDKVEEDLVSDYFSNCGIFSTYWQKYLNFFSTSSCSFQSNIIE ANLSSSLKDWLHELKQLIKEHYEKYWLPENMNLYIDSALDYRLFEKLLASEMATFSSECD NKGKLFIENDLFMQEIEIKKKYGRKLEESKNQGFDENVDLNDRKSNKTIKIFNVVPLSYK KSVLDIDYKIKLKNISQLVSISLTRVMDVVTTLFLDSKMVKRLRDELGIIRDLKLTWRLH YSSLKLRLIFRFILKSMEVSNARKVMEDFYSYSIFLFRQFERTNGRKKSNPEFKVSKELF DIVDEIQRLNEINWHLQYLYEDLPTQLTTPINNCGTPIQVSQPNNLESNWIKNEIRIYNP LLFKNYPQLILSSIHRSLTRLPKECDIMKRKKCNCYRTDHSGKISKANSILDTGSIVFVL MEFIIKKIVELKANVIFTDPYFRYRTDFVLKVRAYNYHEYVNLQFSSYYQKNNLLKEDFT DFTNSENWTLPERIVNFETSMLIPDYEYYKNSTAINRQLSYPRISYKNELSLTTRLENFK FYSPIVFVRSIIRTKLSPCKIFKDCFCEVDCISKRHTGLIMLKILSDIFNISIEKFLESE PIPGYMNILEKLSHNSSTKYGNWRISDSLTKPFHFGSIMIGNNEIELNFVGPSCSLIEYM RKIFKEMNSRFKPLKIVKFYQILRNFMKNEIKRRKNRTPMDFIRYYQGILQYEDYFEDES FITTGISITYDNYLSFHNYIIDTLFNKDSDRPRQKGFMETVVLGITSDEFNKRYTETILD WIYFEKPDSYKEICMLQEPYFAFSAGDEPLIIRQTFTDGILTTSSASIRFQLPCNKNSST GFNSSLSDNDIFFKDEKEYKWETNDGLQPRCIFNTVASRVLERILRKYFSSIVRKLVLEM SQSHSNNTNLPPNLNSNFAVRGEVEVSFYRLNSIPQFTLYILSSHFDSFTLLSILNSSIK RFKTDIIDSEEVLSEEEFSSVKSYFIRYYCIKEKIQIRNLRASLLEMSKWRYNYNWKHES CNLIRTLKKKHIVQFYYDYFLMDSPYRRSYILLLQAPPFNRLISYSKQKGTSIDPVSTVF QNYIVSLLGKCDKADYYRQTNTTKDADWNISMLSYPRILQSDIGILDYKYKNIKKYQPRC SKFMLEAINTEKQN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd7_770 | 366 S | NLSSSLKDW | 0.997 | unsp | cgd7_770 | 366 S | NLSSSLKDW | 0.997 | unsp | cgd7_770 | 366 S | NLSSSLKDW | 0.997 | unsp | cgd7_770 | 465 S | NDRKSNKTI | 0.995 | unsp | cgd7_770 | 709 S | SKANSILDT | 0.996 | unsp | cgd7_770 | 820 S | NRQLSYPRI | 0.994 | unsp | cgd7_770 | 825 S | YPRISYKNE | 0.997 | unsp | cgd7_770 | 1089 S | EKPDSYKEI | 0.998 | unsp | cgd7_770 | 1104 S | YFAFSAGDE | 0.993 | unsp | cgd7_770 | 1127 S | TSSASIRFQ | 0.992 | unsp | cgd7_770 | 1145 S | GFNSSLSDN | 0.994 | unsp | cgd7_770 | 1147 S | NSSLSDNDI | 0.994 | unsp | cgd7_770 | 1274 S | EEVLSEEEF | 0.996 | unsp | cgd7_770 | 1303 S | NLRASLLEM | 0.993 | unsp | cgd7_770 | 1364 S | NRLISYSKQ | 0.99 | unsp | cgd7_770 | 50 S | TLYYSPIEG | 0.994 | unsp | cgd7_770 | 307 S | DPKKSKEEE | 0.998 | unsp |