

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd8_3200 | OTHER | 0.999849 | 0.000103 | 0.000048 |
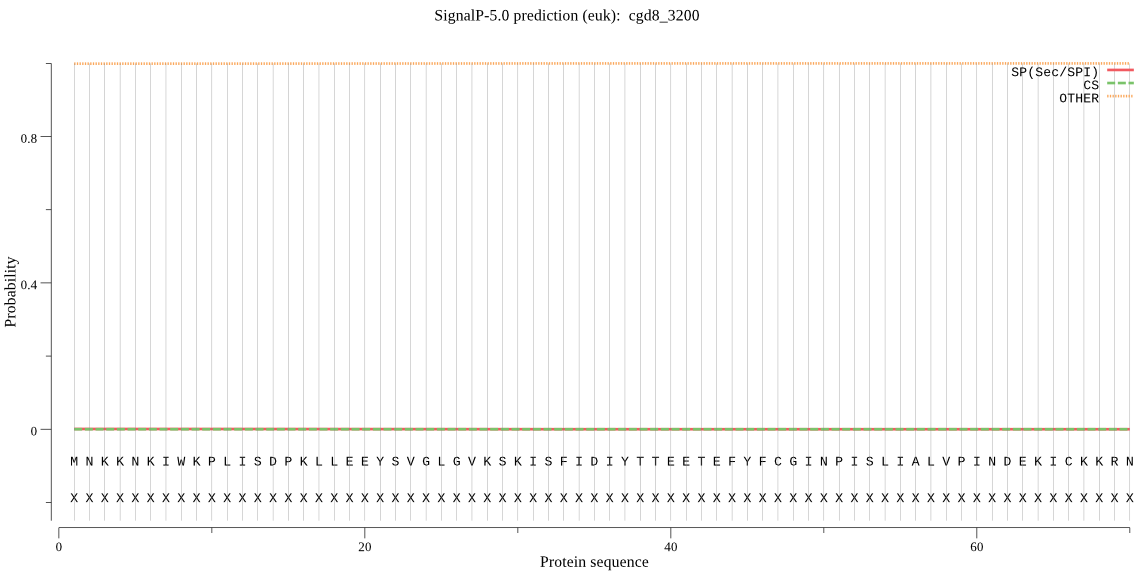
MNKKNKIWKPLISDPKLLEEYSVGLGVKSKISFIDIYTTEETEFYFCGINPISLIALVPI NDEKICKKRNKLGCEMNISQSVWFMKQYITNSCSAVALLHSILNNDKIELEEESIAKMLL NLKGDPNDLPEERGFYLINDKNIEYLHEELSSRDLTKDCDQSEFHYVSFVNNHGHIIELD GRLPCPISHGVCKSDEFLKNTLKIIKENFILPIGINGKIAIVALCIK