|
Algorithm |
|
Prediction of bacterial virulent protein sequences has
implications for identification and characterization of novel
virulence-associated factors, finding novel drugs/vaccine targets against pathogens,
and understanding the complex virulence mechanism in pathogens. The prediction of virulent proteins will aid studies aimed
at knowing more about bacterial virulence and annotation of (unknown)
virulent genes for the identification of novel antimicrobial targets. Hence,
in the present study, an attempt has been made to develop reliable SVM-based
method for the automated prediction of virulent proteins. The training was
carried out using non-redundant dataset of virulent and non-virulent proteins
and evaluation with 5-fold cross-validation technique. In addition
independent datsets were also used for evaluating
the unbiassed performace
of different SVM module. User can download the sequences of different
datasets (Training and Independent datasets) by clicking the link DATA Details about CASCADE SVM Sometimes, machine learning techniques are unable to
handle the noise produced due to the large/complex number of input
units/patterns, which further, effects their
classification efficiency. However, this problem can be overcome by the
construction of very important module i.e. cascade SVM. In the present study,
two-layered cascade approach-based SVM module was constructed. The brief
description about each layer is as follows First layer In the first layer 5 modules based on protein features
such as amino acid compositions, dipeptide composition, amino
acids composition of divided protein sequences, PSSM profiles and
similarity-search were developed. These modules gave SVM predicted scores and
similarity-search based information for each sequence. Second layer The second layer received the binary scores output
generated by 5 best modules constructed in the first layer
to train second layer SVM model. Here, SVM was provided with a vector of 7
dimensions (1 for amino acid composition, 1 for dipeptide composition, 1
amino acids composition of divided protein sequences, 1 for PSSM and 3 for
similarity-search based results). Hence, second layer correlates the
predicted information of the first layer models to provide final output as shown in the picture below 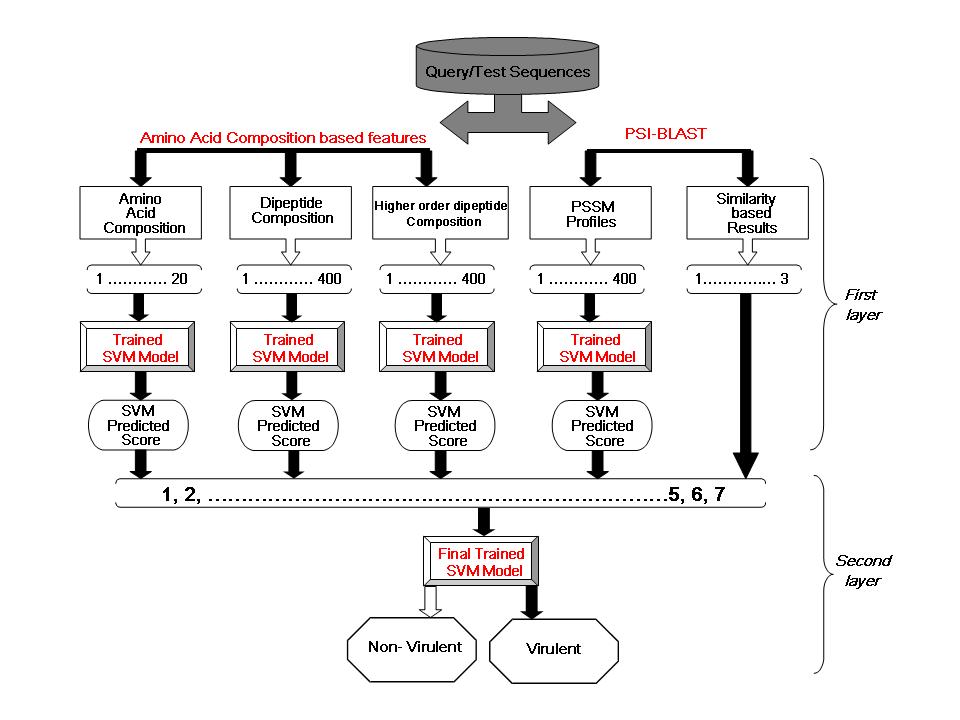 |