Two major search options are available for querying the PAmiRDB database: Search options
- A Comprehensive target search – This is an option where a specific and non-specific search is possible, depending on the requirement of a user.
- Predicted targets of a query miRNA
- Target of miRNAs in a specific virus(can be searched using GenomeID/Virus name)
- Target of miRNAs on a specific viral protein
- Query2 – This option encompasses results at the 10th and 11th position is A/U and compulsory binding. The 10th and 11th positions are very crucial regarding Plant miRNA.
All queries consider individual miRNAs and not miRNA families.
Preloaded search query examples can be found by hovering the mouse on question mark icon near textbox.
Comprehensive target search
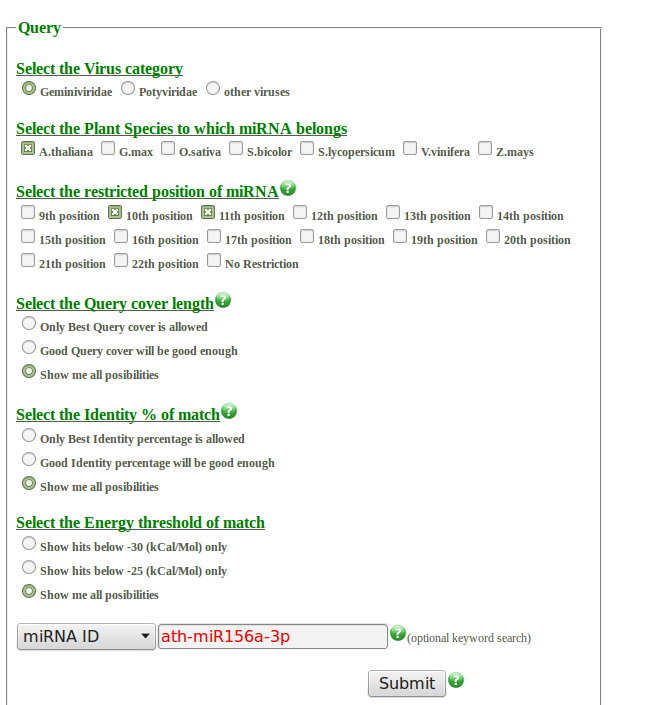
- Select the virus category: By clicking on a radio button, one of the three categories can be selected. The default option is geminiviridae.
- Geminiviridae: 336 geminivirus genomes can be searched against plant miRNAs in order to find interactions.
- Potyviridae: 138 potyviridae genomes can be searched against plant miRNAs in order to find interactions.
- Other virus: This option is comprised of 25 viruses, which don't belong to Potyviridae and geminiviridae families
- Select the Plant Species to which miRNA belongs: By clicking on the checkbox, combinations or individual plant species out of the seven categories can be selected. The default option is A.thaliana. Number of miRNA used per species are listed below.
- A.thaliana: 427 miRNA
- G.max: 639 miRNA
- O.sativa: 713 miRNA
- S.bicolor: 241 miRNA
- S.licopersicum: 110 miRNA
- V.vinifera: 186 miRNA
- Z.mays: 321 miRNA
- Select the restricted position of miRNA: This option gives an opportunity to select compulsory binding positions in result alignment. This is an inclusive option e.g. This is an inclusive search option i.e. if checkbox 10th position and 11th position is selected, it will show all the hits with compulsory binding at position 10 and position 11 from 5' end of miRNA irrespective of binding status at other positions. Seed region(2-8) have compulsory binding in all results. The default option is compulsory binding at the 10th and 11th position. If none of the checkboxes is clicked then all possible results will be displayed.
- Select the Query cover length: This option indicate towards the length of alignment
- Only Best Query cover is allowed: Results with alignment length 18 nucleotides or more will display only.
- Good Query cover will be good enough: Results with alignment length 16 nucleotides or more will display only.
- Show me all possibilities: This radio button will display all the results irrespective of query cover.
- Select the Identity % of the match: This option indicate towards the percentage of Watson and Crick pairing in alignment
- Only Best Identity percentage is allowed: Results with 80% or more Watson and Crick pairing in alignment will be displayed only.
- Good Identity percentage will be good enough: Results with 70% or more Watson and Crick pairing in alignment will be displayed only.
- Show me all possibilities: This radio button will display all the results.
- Select the Energy threshold of the match: This option indicates towards the maximum energy of alignment allowed.
- Show hits below -30 (kCal/Mol) only: Results with Energy below -30 (kCal/Mol) will be displayed only.
- Show hits below -25 (kCal/Mol) only: Results with Energy below -25 (kCal/Mol) only will be displayed only.
- Show me all possibilities: This radio button will display all the results.
- Drop-down menu and keyword search: This option give an opportunity of keyword search.
- miRNA ID: Select the option miRNA ID and type desired miRNA ID in the text box for specific results. e.g. ath-miR156a-3p (specifically for A.thaliana miRNA) or miR156 for any species.
- Genome ID: By selection of this option all the results in the database related to a specific virus can be searched. e.g NC_004090.1
- Name of the virus: In this option rather than giving GenomeId, name of the virus can be used as the query which will result in the display of all the hits from the database with the related virus.
- Protein: Select this option and type the name of a target protein in the textbox, now only those results will be displayed in which target protein is the one, which is searched by the user.
- Energy: Select this option and type the Energy threshold, It must be a negative value and lower than or equal to -20 (kCal/Mol) moreover energy must be in two digits only and not accompanied with units. eg. -20
*If search box if left blank then query will result in all the records meeting specified conditions
Query2
This query shows result where at the 10th and 11th position is A/U and compulsory binding, Query cover is at least up to 18th Nucleotide, mismatches in alignment are 2 or below, Identity > 80.00%.
Result of Comprehensive Search
With default options result page will look like following:
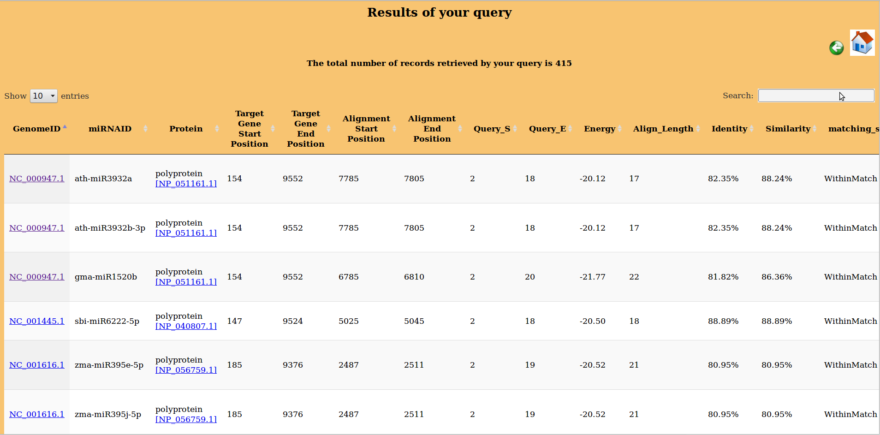
Detail of result columns is as under:
- Virus GenomeID: By clicking on this link all results related to this genome Id corresponding to species of miRNA will be displayed along with alignment details. e.g in the first row click on NC_000869.1 results into all results related to interactions of NC_000869.1 with miRNA from A.thaliana. Specific result related to particular miRNA on that page can be searched using control+F.
- Virus_name: This column refer to the name of virus and ID given along with name direct to NCBI taxonomic browser page related to the same virus on clicking
- Host: This column refer to the name of host of corresponding and ID given along with host name direct to NCBI taxonomic browser page related to the same host on clicking
- miRNAID: This is the miRNAID of specific miRNA about whom all other details are given in a corresponding row.
- Protein: This is the name of Viral protein which is coded by gene where miRNA binds to the viral genome.
- Energy(kCal/Mol): This column indicates this binding energy of interaction between miRNA and viral gene.
- Query_S: This is the starting nucleotide of miRNA
- Query_E: This is the last nucleotide of miRNA involved in alignment. So this column also indicates query cover i.e. number of miRNA nucleotides involved in alignment.
- Target gene begin_position: This is the starting position/nucleotide of the target gene in the viral genome.
- Target gene end_position: This is the last position/nucleotide of the target gene in the viral genome.
- Alignment Start Position: This is the starting position/nucleotide of the viral genome where miRNA binds.
- Alignment End Position: This is the last position/nucleotide of the viral genome where alignment with miRNA ends.
- Alignment Length: This is the number of viral nucleotide and gaps involved in alignment.
- Identity: This column indicates the percentage of Watson and Crick base pairing in alignment.
- Similarity: This column indicates the percentage of Watson and Crick base pairing along with wobble base pairing (GT/GU) in alignment.
- Matching status: Withinmatch- The interaction of miRNA with viral genome is within a single gene. Overlapping- This signifies this condition where miRNA didn't bind within a single gene.
- Target gene details: This column gives further details about target gene.
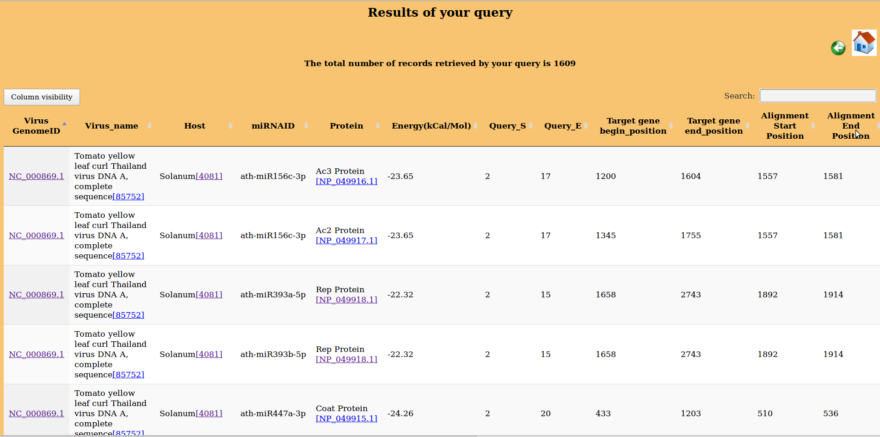
Result of Query2
When checkbox against complete record is checked, result page will look like as under: