

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_082600 | SP | 0.033559 | 0.965860 | 0.000581 | CS pos: 16-17. SLS-KQ. Pr: 0.5992 |
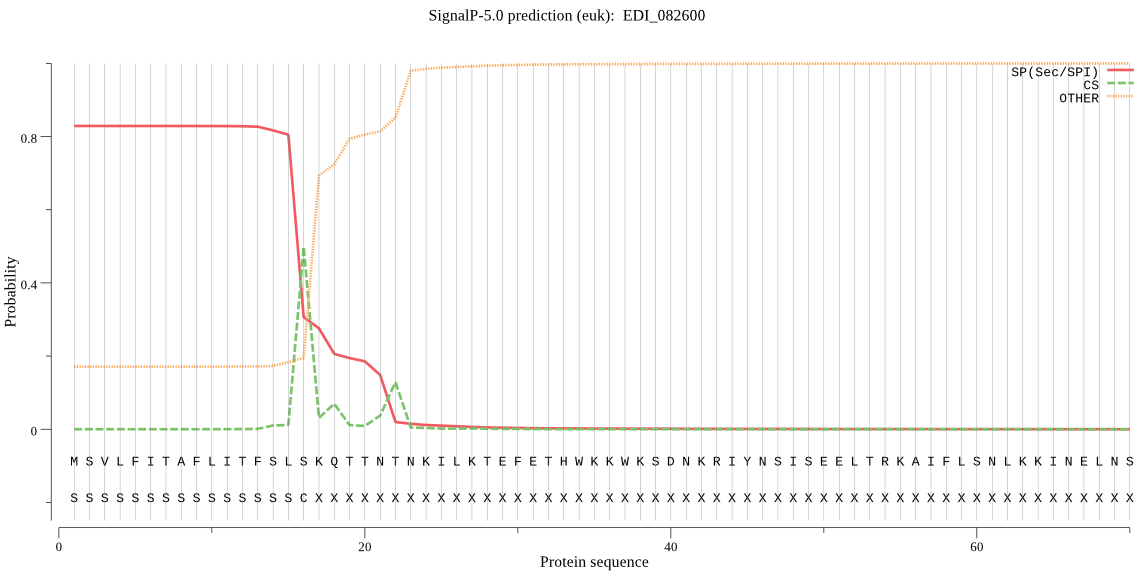
MSVLFITAFLITFSLSKQTTNTNKILKTEFETHWKKWKSDNKRIYNSISEELTRKAIFLS NLKKINELNSQRIDPDDAVFGLNAFSDLKPEEFSRRFNKINLKLLKLKQRNHYKLPVPSG EVPTQYSACLQDMLLDKNSSNEIDLCGGIVIDQGDCGSCYAISNAHELQLKYANLTLTQR GKIEYEMFSPQQLMDCTENSYYCEGGTADEPLMSSHYVVFEKDYPYISYSNTLINNSCVH DKLTPMKVSFSLFDSAQNFEILKRITYYYGSFVTSVKASSDWSYYHSGIYSHSCTKNVVT NHVIEIVGYGNHNGKEYLIARNSWGKNWGIDGFIKISAKSLCGIGGDDGGIYPVSLIQHT DFSDVPKGKYGKKSYKQEVQPKLYNGTDKYDIYEQFPDSSGDSFIDIIISFIVTIPLVVG LISWAICAIVILILSCVFYLNKN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_082600 | 399 S | QFPDSSGDS | 0.997 | unsp | EDI_082600 | 399 S | QFPDSSGDS | 0.997 | unsp | EDI_082600 | 399 S | QFPDSSGDS | 0.997 | unsp | EDI_082600 | 403 S | SSGDSFIDI | 0.994 | unsp | EDI_082600 | 251 S | KVSFSLFDS | 0.991 | unsp | EDI_082600 | 374 S | YGKKSYKQE | 0.996 | unsp |