

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
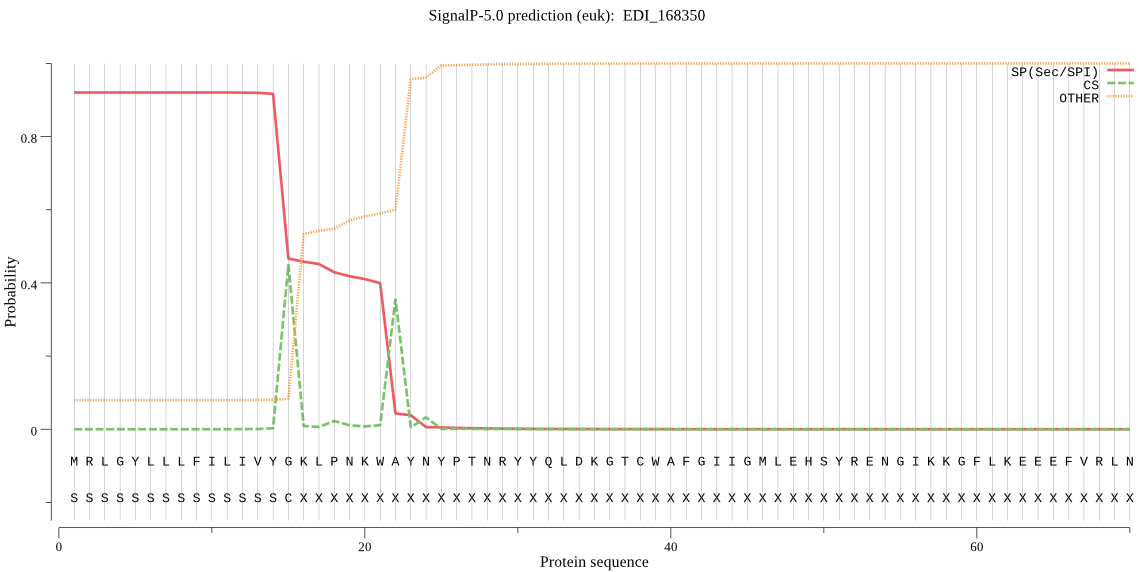
| EDI_168350 | SP | 0.006200 | 0.993756 | 0.000044 | CS pos: 22-23. KWA-YN. Pr: 0.6085 |
MRLGYLLLFILIVYGKLPNKWAYNYPTNRYYQLDKGTCWAFGIIGMLEHSYRENGIKKGF LKEEEFVRLNVQSFGILMVDACKKYPSVCNTPGDDVIFGSTEGGEINWFYSFPFLYDKIL PSAVCPYTATVDTQFECNGMDEALKTNPIKFNITEMLTTYNEEQTKELLLKVKIPIGFGA LIHDAKYYLPCTEEYKNFCDESVYNVIECPENMKYLAEKCAYIVMPMYSTDGEFNYHNEI EPEGGHAMVTVGYNDEYVTHEGCKGGFILKNSWNDTVYGPSIANTARGVRGSHSVKYFMN QLTAEEERKVCPNAQDPMNWYICDDACVINEELHKTIVNELYQAYKLQCVNPEEHFCETG YDYYLTELKADSKSPMNHYYIATFTKYDSTGKKLDTITLPSLPTSIIGMIFTPVEDQLIR LHDSEEFCGHYMFPYCILNKHLPFWGGYVGSHFEIEWDDRSYLINKDKYPEFDYKFIAQS TFVQNLKLVDQKAGVPFLNERI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_168350 | 374 S | ADSKSPMNH | 0.991 | unsp | EDI_168350 | 374 S | ADSKSPMNH | 0.991 | unsp | EDI_168350 | 374 S | ADSKSPMNH | 0.991 | unsp | EDI_168350 | 50 S | MLEHSYREN | 0.993 | unsp | EDI_168350 | 272 S | ILKNSWNDT | 0.992 | unsp |