

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
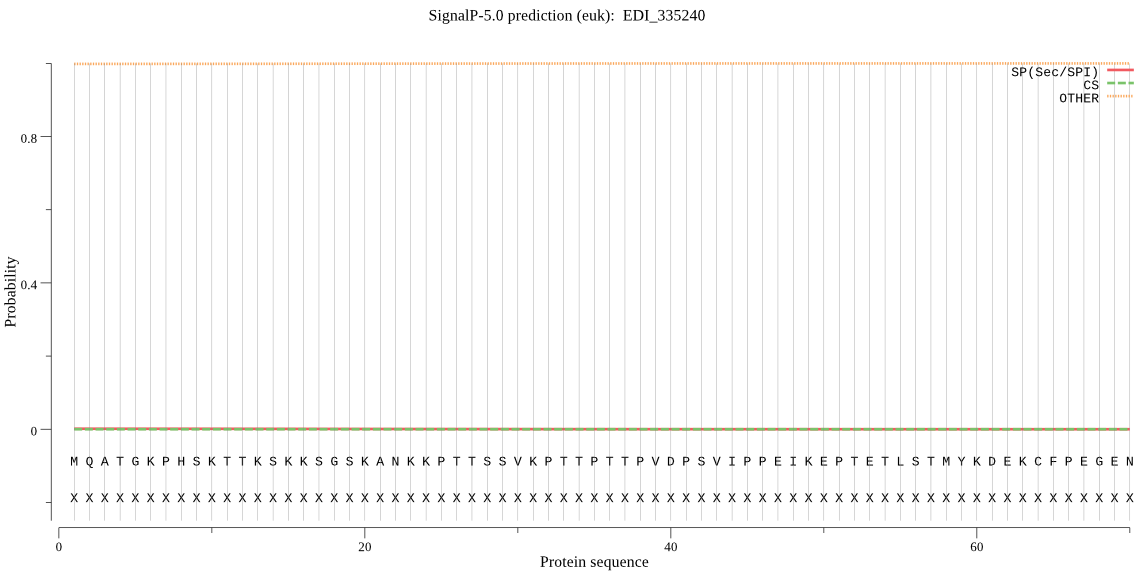
| EDI_335240 | OTHER | 0.999968 | 0.000007 | 0.000025 |
MQATGKPHSKTTKSKKSGSKANKKPTTSSVKPTTPTTPVDPSVIPPEIKEPTETLSTMYK DEKCFPEGENMQYTQEWNLQRFTSAEARERERALIDIEDYRKAAAIHKSVRQWAQQWIKP GMSDLFVAENIERKVREECEVDGVNTTERGMAFPCGLSVNSCAAHFTPNPNDPLSFYKTD DVVKIDFGVHVNGHLIDSAFTMTWDPTLQPILDCAKDATNTGIKNIGVDVRLCDIGDAIE EVMNSYEVEIKGKTYQLQPVRNLSGHMVGSYAVHAGKSIPICKGGPQTKMEEGEVYALET FATTGRGRVDDSGPASHYMVDANAFDYPVRDGNAKRLLRALDANFKTLAFCRRYVDKIGF AKWQMPFKFLVDDGCVNPYPPLSDCHGSYVAQFEHTIYLKPTCKEVLSRSFDY
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_335240 | T | 33 | 0.712 | 0.650 | EDI_335240 | T | 37 | 0.712 | 0.146 | EDI_335240 | T | 26 | 0.707 | 0.117 | EDI_335240 | T | 27 | 0.705 | 0.186 | EDI_335240 | T | 36 | 0.705 | 0.697 | EDI_335240 | T | 34 | 0.701 | 0.256 | EDI_335240 | S | 42 | 0.651 | 0.500 | EDI_335240 | S | 29 | 0.599 | 0.253 | EDI_335240 | T | 4 | 0.592 | 0.431 | EDI_335240 | S | 28 | 0.590 | 0.070 | EDI_335240 | T | 52 | 0.541 | 0.074 | EDI_335240 | T | 12 | 0.527 | 0.031 | EDI_335240 | S | 19 | 0.525 | 0.028 | EDI_335240 | T | 11 | 0.510 | 0.026 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_335240 | T | 33 | 0.712 | 0.650 | EDI_335240 | T | 37 | 0.712 | 0.146 | EDI_335240 | T | 26 | 0.707 | 0.117 | EDI_335240 | T | 27 | 0.705 | 0.186 | EDI_335240 | T | 36 | 0.705 | 0.697 | EDI_335240 | T | 34 | 0.701 | 0.256 | EDI_335240 | S | 42 | 0.651 | 0.500 | EDI_335240 | S | 29 | 0.599 | 0.253 | EDI_335240 | T | 4 | 0.592 | 0.431 | EDI_335240 | S | 28 | 0.590 | 0.070 | EDI_335240 | T | 52 | 0.541 | 0.074 | EDI_335240 | T | 12 | 0.527 | 0.031 | EDI_335240 | S | 19 | 0.525 | 0.028 | EDI_335240 | T | 11 | 0.510 | 0.026 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_335240 | 14 S | KTTKSKKSG | 0.995 | unsp |