

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
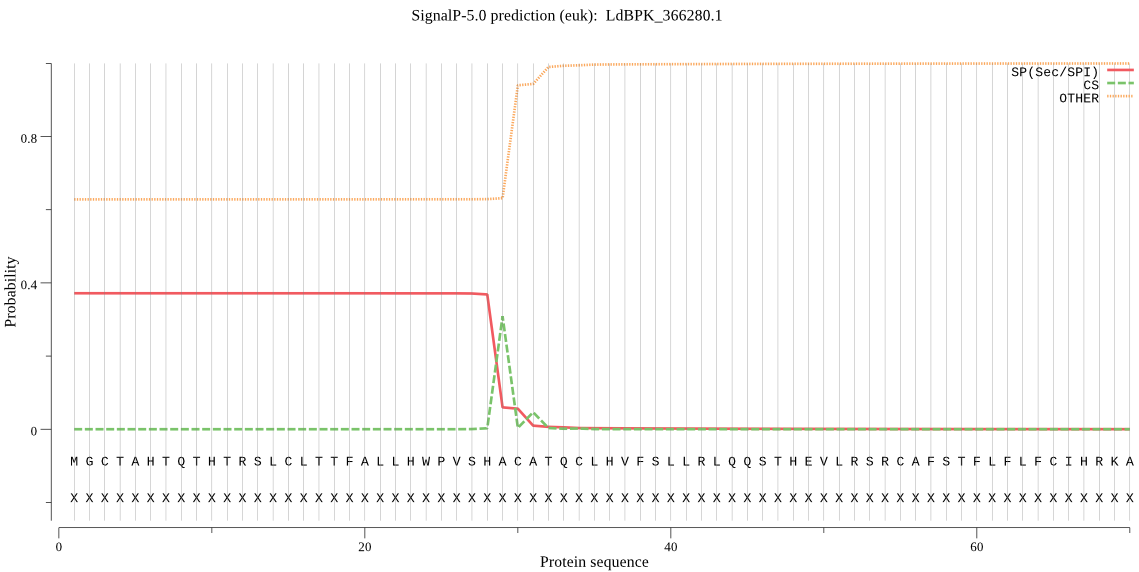
| LdBPK_366280.1 | SP | 0.260073 | 0.738537 | 0.001391 | CS pos: 29-30. SHA-CA. Pr: 0.8223 |
MGCTAHTQTHTRSLCLTTFALLHWPVSHACATQCLHVFSLLRLQQSTHEVLRSRCAFSTF LFLFCIHRKASVPQPLTALSCGRCAPTVEPRFSVFSHRPANTQTHTQTQTERHLPCIMSI SVRIRTPKSSTPEQVELNSSGTWGEAAALLSRKSEVALERLRVLAGFPPKAVDLAADSPL SALKLRANDMLIVQEGEAKVQLGNRGERYLPPAPERAHLTRRRCPADNSCLFHACAYVLR DKSRTEGPQLRQECVQAVLDHPEMFNVNTLGMDPLAYASWLSEKDTWGGAIELEILSFLY KTEIFALDLQSVTLQRFGTGMGYTVRAFLVYTGNHYDCIAMNPVYNPTSEREDQTLFSSR DENVLARAKRFVAEEGQKMKEGKST
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_366280.1 | 282 S | ASWLSEKDT | 0.993 | unsp | LdBPK_366280.1 | 282 S | ASWLSEKDT | 0.993 | unsp | LdBPK_366280.1 | 282 S | ASWLSEKDT | 0.993 | unsp | LdBPK_366280.1 | 358 S | QTLFSSRDE | 0.99 | unsp | LdBPK_366280.1 | 130 S | TPKSSTPEQ | 0.995 | unsp | LdBPK_366280.1 | 243 S | LRDKSRTEG | 0.997 | unsp |