

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
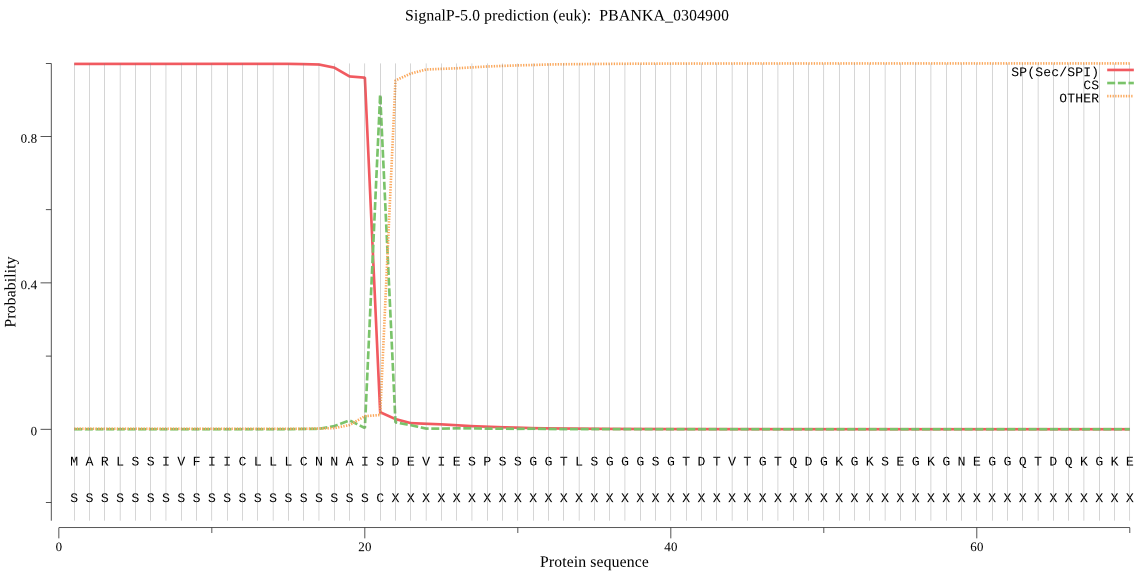
| PBANKA_0304900 | SP | 0.000788 | 0.999212 | 0.000000 | CS pos: 21-22. AIS-DE. Pr: 0.9650 |
MARLSSIVFIICLLLCNNAISDEVIESPSSGGTLSGGGSGTDTVTGTQDGKGKSEGKGNE GGQTDQKGKENPENGQNSDSTGDSSLGSTGSNGSQPAPTTPKEPEPTTPKEPESATPKAS EPVTPQKTAETASGKQVSPTPSENPPSKDTPKPESSSEKKVNSALATPPAPEVSKAQEGA GLATQKEQTPSKRAKRSPPPQVNNITDMESYLMKNYDGVKVIGLCGVYFRVQFSPHLLLY GLTTFSIIQIEPFFEGVRIDFEHQHPIRNKCAPGKAFAFISYVKDNILILKWKVFAPPSD LFANDEVSKQILSAVSVTSDAESSVVDVRKYRLPQLDRPFTSIQVYKANPKQGLLETKNY ILKNAIPEKCSKISMNCFLNGNVNIENCFKCTLLVQNAKPTDECFQYLPSDMKNNLNEIK VTAQSDEDSKENDLIESIEILLNSFYKADKKAKKLSLITMDDFDDVLRAELFNYCKLLKE LDTKKTLENAELGNEIDIFNNLLRLLKTNEEESKHNLYKKLRNTAICLKDVNKWAEKKRG LILPEEVTQDQMAIGQNEEPYDEDPDDRVDLLELFDDNQNENIVDKDGIIDMSIAIKYAK LKSPYFNSSKYCNYEYCDRWQDKTSCISNIDVEEQGNCSLCWLFASKLHLETIRCMRGYG HNRSSALYVANCSKRTAEEICNDGSNPLEFLKILEKNKFLPLESNYPYLWKNVSGKCPNP QNDWTNLWGNTKLLYNNMFGQFIKHRGYIVYSSRFFAKNMNVFIDIIKREIRNKGSVIAY IKTQGVIDYDFNGRYISNICGHNHPDHAVNIIGYGNYISESGEKRSYWLIRNSWGYYWGH EGNFKVDVLGPDNCVHNVIHTAIVFKIDMEPDSDSNNNNAIKNRDQLIDEDNKSYFPQLS SNFYHSLYYNNYEGYEAKNDDENDQNYGNLDVSGQSENHQNDPKNPQPQANSTVTQLGVC PAENQRTADSNPNAQSTPSPNTTVTDTVNSNTANSNTANSNTASANAASIEKKIQILHVL KHIEKYKMTRGFVKYDNLNDTKNDYTCARSYSYDPKNHNECKQFCEENWERCKNHYSPGY CLTTLSGKNKCLFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_0304900 | 100 T | PAPTTPKEP | 0.996 | unsp | PBANKA_0304900 | 100 T | PAPTTPKEP | 0.996 | unsp | PBANKA_0304900 | 100 T | PAPTTPKEP | 0.996 | unsp | PBANKA_0304900 | 108 T | PEPTTPKEP | 0.995 | unsp | PBANKA_0304900 | 138 S | GKQVSPTPS | 0.991 | unsp | PBANKA_0304900 | 150 T | PSKDTPKPE | 0.993 | unsp | PBANKA_0304900 | 155 S | PKPESSSEK | 0.997 | unsp | PBANKA_0304900 | 425 S | VTAQSDEDS | 0.998 | unsp | PBANKA_0304900 | 39 S | SGGGSGTDT | 0.991 | unsp | PBANKA_0304900 | 80 S | QNSDSTGDS | 0.997 | unsp |