

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
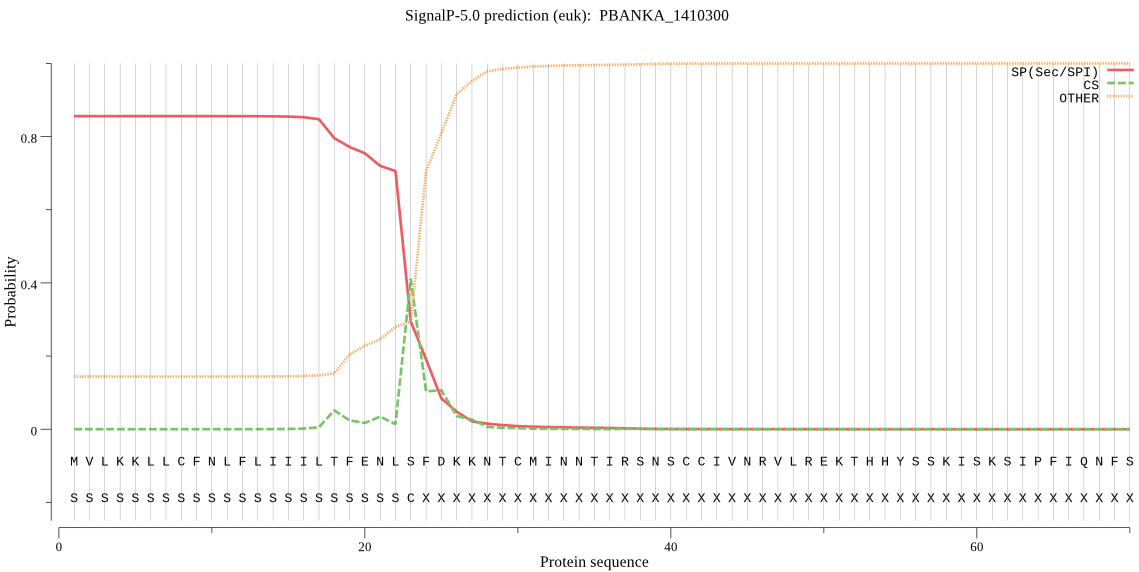
| PBANKA_1410300 | SP | 0.058666 | 0.941080 | 0.000255 | CS pos: 23-24. NLS-FD. Pr: 0.3791 |
MVLKKLLCFNLFLIIILTFENLSFDKKNTCMINNTIRSNSCCIVNRVLREKTHHYSSKIS KSIPFIQNFSLEKYFTGESLQKNILNNINKLGGAHLFHISKSHLTAKSGNKNTEFIGEAT ELFKGFKRNFGINMTENKQTNIGRMLCENDNNNGGEDTSTEKAIFKKSKDSQIHYRTDYK PSGFTIDNVTLNINIFDNETIVRSSLNMCTNENYADEDLVFDGVGLSIKEISINNNKLTE GEDYTYDNEFLTIFAKNVPKENFVFLSEVVIHPETNYALTGLYKSKDIIVSQCEATGFRR ITFFIDRPDMMAKYDVTLTADKKKYPVLLSNGDKLNEFDIPGGRHGARFNDPHLKPCYLF AVVAGDLKHLSDNYVTKYTKKPVELYVYSEAKYVSKLKWALECLKKAMKFDEDYFGLEYD LSRLNLVAVSDFNVGAMENKGLNIFNADSLLASKKTSIDFSFERILTVVGHEYFHNYTGN RVTLRDWFQLTLKEGLTVHRENLFSEETTKTATFRLTHIDLLRSVQFLEDSSPLSHPIRP ESYISMENFYTNTVYDKGSEVMRMYQTILGDDYYKKGIDIYLKKHDGGTATCEDFNDAMN EAYQMKKGNTDENLDQYLLWFSQSGTPHVTAEYIYDENEKTFTINLSQITYPDDNQKEKY PLFIPVKVGFISPKDGKDVIPETVLELKKDKESFVFQNVSEKPIPSLFREFSAPVYIKDN LTDEERIALLKYDSDAFVRYNVCIDLYMKQIIKNYNELISQKTKENNVLELSLTPVNDEF INAIKHLLEDKHADPGFKSYIIALPRDRYIMNYIKEVDPIVLADTKDYIYKQIGSRLNPV LFSIFQNTESKANDMTHFKDESYIDFDQLNMRKLRNSILMMLSKAQYPHMLKYIKEQSNS PYPSNWLASLSASSYFSGDDYYDLYDKTYKLSKNDELLLQEWLKTVSRSDRSDIYSIIKK LEVEILKDSKNPNNIRAVYLPFTSNLRAFNDISGKGYKLMANVIMKVDKFNPMVATQLCD PFKLWNKLDLKRQALMHDEMNRMLNMENISPNLKEYLLRLTNKM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1410300 | 672 S | VGFISPKDG | 0.998 | unsp | PBANKA_1410300 | 672 S | VGFISPKDG | 0.998 | unsp | PBANKA_1410300 | 672 S | VGFISPKDG | 0.998 | unsp | PBANKA_1410300 | 917 S | SSYFSGDDY | 0.993 | unsp | PBANKA_1410300 | 227 S | GVGLSIKEI | 0.991 | unsp | PBANKA_1410300 | 542 S | IRPESYISM | 0.992 | unsp |