

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
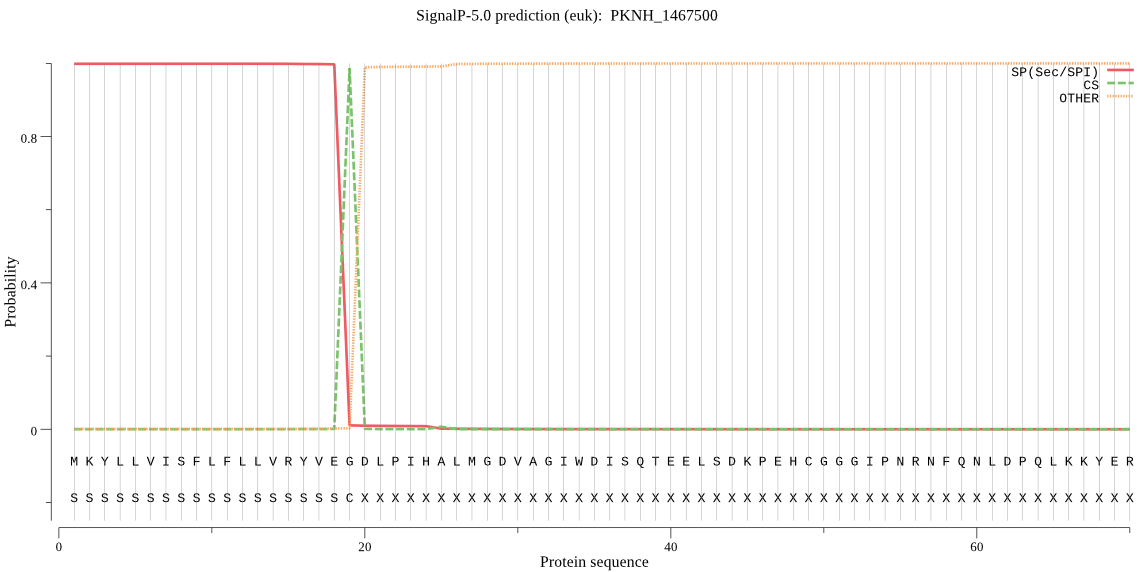
| PKNH_1467500 | SP | 0.001203 | 0.998776 | 0.000020 | CS pos: 19-20. VEG-DL. Pr: 0.9885 |
MKYLLVISFLFLLVRYVEGDLPIHALMGDVAGIWDISQTEELSDKPEHCGGGIPNRNFQN LDPQLKKYERFLENNYGGMETTSMKLTTEKINLTDKLNQRNNWTYLAVRDANTNGIIGHW TMVYDEGFEVRVPGRRYFALFKYERTNSKKCPEPIENKDSTDSNCYITDPTRTLLGWVLH ERVHENDKKKKVFQWGCFYGRKQEDVGISSFVIHGAHSSPPSTEEREKSMGSSTWTGGNI LLPTEKQLSFASIKQRSNYTKIRLSSGSPHGVPKTSLMSIYQRTKERIFGCRKEDSEEVK IRLTLPKAFTWGDPFSDDNFEEDVEDQMNCGSCYSIATLYSLQKRFEIGLFKKYRKKITV PKLSYQSILSCSPYNQGCDGGFPFLVGKHLYEFGIPAEDSFPYGMSDSIKCEMSMGSYNN ITSTTYKEKDLFFVGEYNYVSGCYECSNEFDMMKEIYSNGPIVVAINATAQLLGLYKLGK QNSMYDIATHENKVCDIPNEGFNGWQQTNHAVTIVGWGEVEKEQEGGNLIKYWVVRNTWG KTWGYKGYIKFQRGVNLAGIETQAVFLDPDLSRGRAGNISGEER
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1467500 | 229 S | EREKSMGSS | 0.993 | unsp | PKNH_1467500 | 229 S | EREKSMGSS | 0.993 | unsp | PKNH_1467500 | 229 S | EREKSMGSS | 0.993 | unsp | PKNH_1467500 | 265 S | KIRLSSGSP | 0.994 | unsp | PKNH_1467500 | 148 S | ERTNSKKCP | 0.996 | unsp | PKNH_1467500 | 222 S | SSPPSTEER | 0.996 | unsp |