

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
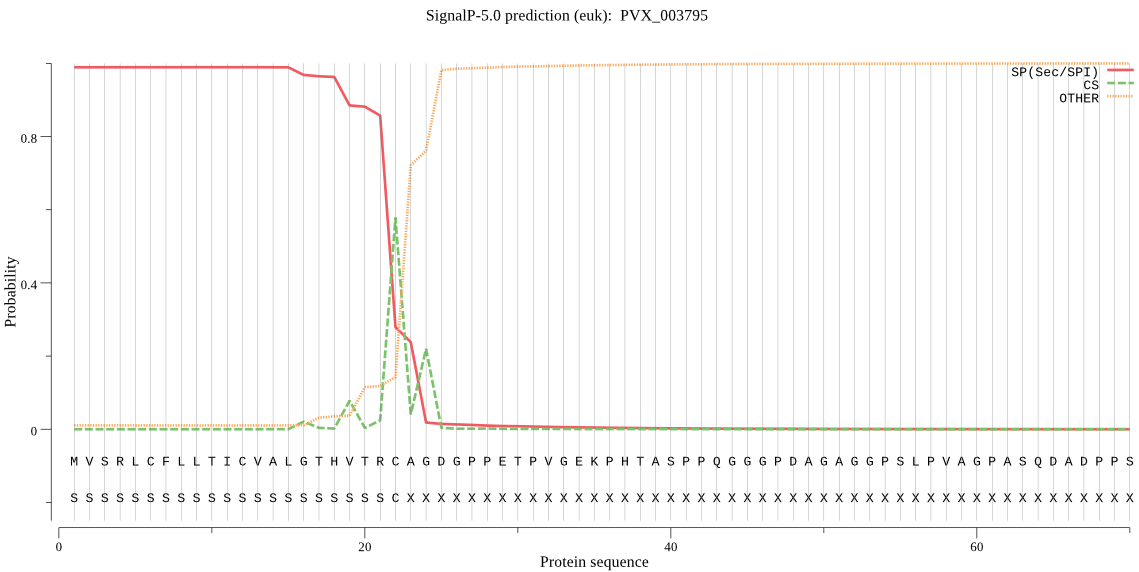
| PVX_003795 | SP | 0.000283 | 0.999713 | 0.000004 | CS pos: 22-23. TRC-AG. Pr: 0.6752 |
MVSRLCFLLTICVALGTHVTRCAGDGPPETPVGEKPHTASPPQGGGPDAGAGGPSLPVAG PASQDADPPSSSSANGIASPPADGETALPPRSAETTDAADVTKSPSSATEPESAGAASTQ AEDDAEQGIQITSALLKHFNGIKITGSCKEPFRVFLSPHLWIYAVPPENKIQLRPNFGPI TSIELGNLSNDCKKGANNSFKLIVHTSDDILTLKWKVTGEGAAPGNKADVKKYKLPTLER PFTSVQVHSANAKSKIIESKFYDIGSGMPAQCSAIATNCFLSGSLEIEHCYHCTLLEKKL AQDSECFKYVSSEAKELIEKDTPIKAQEEDANSADHKLIESIDVILKAVYKSDKDEEKKE LITPEEVDENLKKELANYCTLLKEVDTSGTLNNHQMANEEETFRNLTRLLRMHSEENVVT LQDKLRNAAICIKHIDKWILNKRGLTLPEEGYPSEGYPPEEYPPEELLKEIEKEKSALND EAFAKDTNGVIHLDKPPNEMKFKSPYFKKSKYCNNEYCDRWKDKTSCMSNIEVEEQGDCG LCWIFASKLHLETIRCMRGYGHFRSSALFVANCSKRKPEDRCNVGSNPTEFLQIVKDTGF LPLESDLPYSYSDAGNSCPNKRNKWTNLWGDTKLLYHKRPNQFAQTLGYVSYESSRFEHS IDLFIDILKREIQNKGSVIIYIKTNNVIDYDFNGRVVHSLCGHKDADHAANLIGYGNYIS AGGEKRSYWIVRNSWGYYWGDEGNFKVDMYGPEGCKRNFIHTAVVFKIDLGIVEVPKKDE GSIYSYFVQYVPNFLHSLFYVSYGKGADKGAAVVTGQAGGAVVTGQTETPTPEAAKNGDQ PGAQGSEAEVAEGGQAGNEAPGGLQESAVSSQTSEVTPQSSITAPQIGAVAPQIGAAAPQ IDVAAPQIDVVAPQTRSVDAPQTSSVAAHPPNVTPQNVTLGEGQHAGGVGSLIPADNSGE KNIPVMHLLKYVKETKMTRAFVTYGSLSEIEGSHSCSRIYAKDEEGHEECAQFCLKNWPA CRGHYSPGYCLTKMYTGSNCLFCSV
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003795 | T | 102 | 0.605 | 0.457 | PVX_003795 | T | 95 | 0.604 | 0.053 | PVX_003795 | T | 38 | 0.602 | 0.089 | PVX_003795 | T | 96 | 0.596 | 0.238 | PVX_003795 | T | 86 | 0.556 | 0.128 | PVX_003795 | T | 109 | 0.534 | 0.213 | PVX_003795 | T | 30 | 0.532 | 0.249 | PVX_003795 | S | 104 | 0.523 | 0.130 | PVX_003795 | T | 119 | 0.507 | 0.398 | PVX_003795 | S | 40 | 0.505 | 0.034 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003795 | T | 102 | 0.605 | 0.457 | PVX_003795 | T | 95 | 0.604 | 0.053 | PVX_003795 | T | 38 | 0.602 | 0.089 | PVX_003795 | T | 96 | 0.596 | 0.238 | PVX_003795 | T | 86 | 0.556 | 0.128 | PVX_003795 | T | 109 | 0.534 | 0.213 | PVX_003795 | T | 30 | 0.532 | 0.249 | PVX_003795 | S | 104 | 0.523 | 0.130 | PVX_003795 | T | 119 | 0.507 | 0.398 | PVX_003795 | S | 40 | 0.505 | 0.034 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003795 | 352 S | AVYKSDKDE | 0.998 | unsp | PVX_003795 | 352 S | AVYKSDKDE | 0.998 | unsp | PVX_003795 | 352 S | AVYKSDKDE | 0.998 | unsp | PVX_003795 | 610 S | DLPYSYSDA | 0.995 | unsp | PVX_003795 | 651 S | LGYVSYESS | 0.995 | unsp | PVX_003795 | 988 S | YGSLSEIEG | 0.996 | unsp | PVX_003795 | 107 S | KSPSSATEP | 0.998 | unsp | PVX_003795 | 147 S | KITGSCKEP | 0.993 | unsp |