

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
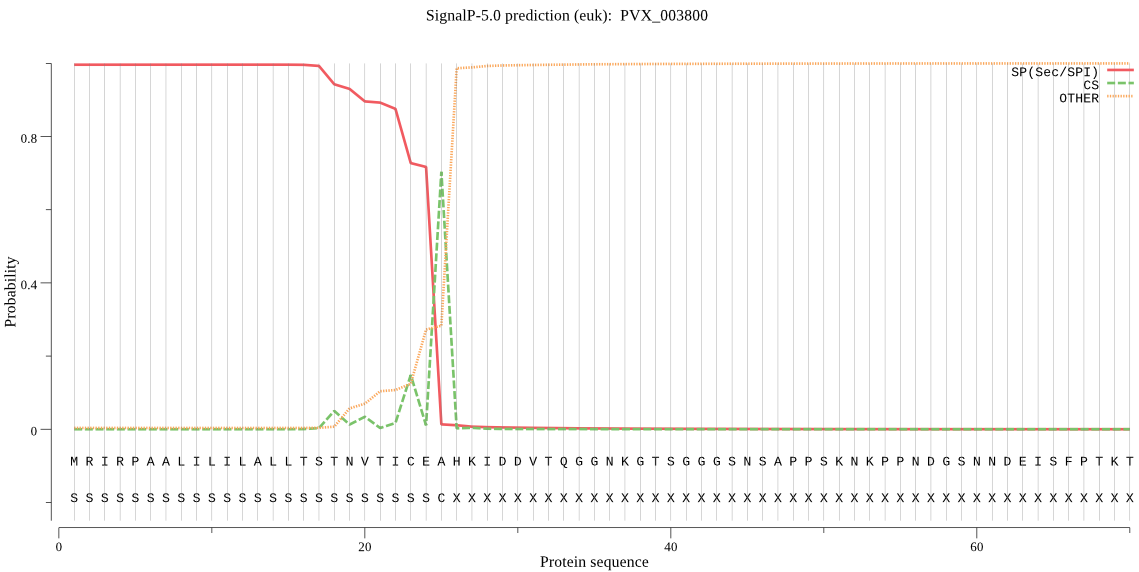
| PVX_003800 | SP | 0.000269 | 0.999731 | 0.000000 | CS pos: 25-26. CEA-HK. Pr: 0.5479 |
MRIRPAALILILALLTSTNVTICEAHKIDDVTQGGNKGTSGGGSNSAPPSKNKPPNDGSN NDEISFPTKTISFSMPPTLEGGGSKNDSAGSADQVGGTTGGSPPSSNNATGGSSNSSGKK AGNTHIPSRGLQKTKSSARGVTQKGHQTAVDIFGGSTTKSNAPHEGPHHVESALLKNYKG VKVTGSCGSYFRVYLVPHILIYALTKNSIIQIESLFDDNTRVDFEYADDIVNKCEEGKNF KLIVYLKGNILTLKWKVLPSSDHSAAGTKADVRKFKLPQLERPFSSIQVYTADVKAGLIE SKNYALGADIPDKCNAIATDCFLNGNVNIDKCFQCALLVQGGDATSNECFNYVSKDLQDK HSNAKVKGQDELSPKEYELTESIDIILKNIYKVDTINKKKVLISMDDFDDVLKAELLNYC KLLKEMDVKGTLDNCELGNEVDIFNNMVRLLSMHPKENIITLQDKLRNTAICLKNVDEWV ENKRGLVLPEGGEKAALEGNPTEVVDDEEEEHAGEGKLLDQDMYKKDEDGTIDLVKAGKE LKLRSPYFKNSKYCNYEYCDRWKDKTSCISNIEVEEQGNCGLCWVFASKLHLETIRCMRG YGHFRSSALYVANCSKRNPKDICTVGSNPIEFLKIVQDTGFLPLEADHPYLHKNAGNECP APKENWINLWGNSKLLFHKMYGHFMSYKGFISYESSHFKDNMHIFIDLIKREVQNKGSVI IYIKTKDVIGYDFNGRGVHNMCGDKTPDHAANIVGYGNYINTKGVKRSYWIVRNSWGYYW GNEGTFKVDMLGPKGCKYNFIHTAVVFKVNLGAIDIPNKDVDLNRTYFPKHNSDFFHSLY FNNYDEEADGEEKPVFQGARSSEEGGSGSGSGSGGSGGGSGTKSKKGSKKKQISGQDAAA ATAGATGGASSIARSGLPPKSPKPSPSAEKKIQILHVLKHIENSKIVRGLVKYENISETQ NDHSCARAHSKNPDRLDECKLFCEENWNSCKNHYSPGYCLAKLYSGGNCYFCYV
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_003800 | T | 110 | 0.501 | 0.062 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_003800 | T | 110 | 0.501 | 0.062 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003800 | 404 S | KVLISMDDF | 0.994 | unsp | PVX_003800 | 404 S | KVLISMDDF | 0.994 | unsp | PVX_003800 | 404 S | KVLISMDDF | 0.994 | unsp | PVX_003800 | 861 S | QGARSSEEG | 0.997 | unsp | PVX_003800 | 862 S | GARSSEEGG | 0.994 | unsp | PVX_003800 | 884 S | SGTKSKKGS | 0.996 | unsp | PVX_003800 | 888 S | SKKGSKKKQ | 0.996 | unsp | PVX_003800 | 894 S | KKQISGQDA | 0.995 | unsp | PVX_003800 | 921 S | LPPKSPKPS | 0.997 | unsp | PVX_003800 | 925 S | SPKPSPSAE | 0.995 | unsp | PVX_003800 | 117 S | SSNSSGKKA | 0.993 | unsp | PVX_003800 | 373 S | QDELSPKEY | 0.998 | unsp |