

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
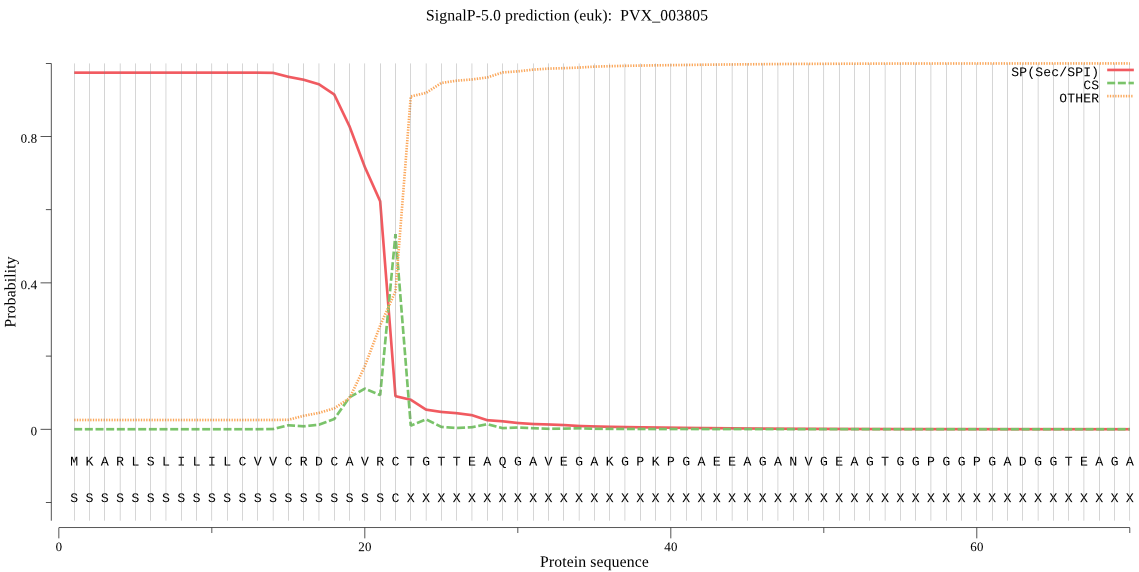
| PVX_003805 | SP | 0.017452 | 0.982388 | 0.000160 | CS pos: 22-23. VRC-TG. Pr: 0.7283 |
MKARLSLILILCVVCRDCAVRCTGTTEAQGAVEGAKGPKPGAEEAGANVGEAGTGGPGGP GADGGTEAGARAEEGEGAGTEAEPEAEPEAEAEPARGPEPEPEAGGEGINRDAAGNQREG QLEAPSDSARPGAIPQVAPRDTVETSSDAADSSSPDQNPLPGADNTKVGNAATPPEGAKE ETQVKSSLLKGHKGVKVTGPCGASFLVFFAPYLFIDVDTDSSNVYLGTDLSDLEVTEKMG IQDNGKNKCEDKKTFKFVALIGEDHLTIKWKVYDPAVKTPTPNEKVEMRKYVMKNLSGEF TAVQVHTVIQQNGSNVFESKNYALSSGIPEKCDAVATNCFLSGSVYIEKCYRCTLKMKKV DPSDVCYNYIPKVESAASQEAIPAKASDEESSQEELTASIGKILQGVYKKGENGLNEVLT FNEADAALKAELLNYCSLMKKVDASGVLGHYQLGSEEDVHANLTNMLQTNSDHVLSSLQN KLKNPAICLKNADEWVGSKTGLLLPNLFYNHLEGSTPSTSNVTHVDDSSEDVQSGGYDGV IDFATAGKTNFSTSQYADKMHCNAEYCDRAKDAGSCVAKMEVQDQGDCANSWLFASKVHL ETIKCVKGYDHVGASALYVANCSGKEANDKCHSPSNPLEFLNTLEETNFLPADSDLPYSY KQVGNACPEPKGHWQNLWENVKLLGPTNEPNSMSTKGYTAYQSDHFKGNMDSFIKLVKSE VMKKGSVIAYVKVAGALSYDLNGKKVLSLCGSETPDLAVNIVGYGNYISAEGVKKPYWLL QNSWGKHWGDKGTFKVDMNGPPGCHHNFIHTAAVFNLDMPVEENPQKEDAQIYNYYLKSS PDFWGNIYYKNVGGQESASSKNATEGAHESVLHGQEVAEAAVNGAGGEPAPSHSGAVQGE REGTANGQGPEGVPPLPAKQEVTEASNGEATGLGAVNPSGEEQPGPPGPPGTSGLAEQPG PEGPSGEVGSTGSVGQPGQPGSSGTPRSEGQTGPSGTSGSSAAQGQSGSSGTPGSDGPSG HTGPSSTPVQEAPLSKAPGTDSPPVAPEAAVLGSEVTHVLKYIKKNKVKLNLVTYKNHEA LSSGHDCWRSYSANPDKYEECVKLCEANWSKCENDAAPGFCLYEHAKEEDCFFCYV
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_003805 | T | 142 | 0.505 | 0.058 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_003805 | T | 142 | 0.505 | 0.058 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003805 | 528 S | HVDDSSEDV | 0.997 | unsp | PVX_003805 | 528 S | HVDDSSEDV | 0.997 | unsp | PVX_003805 | 528 S | HVDDSSEDV | 0.997 | unsp | PVX_003805 | 554 S | NFSTSQYAD | 0.992 | unsp | PVX_003805 | 859 S | QESASSKNA | 0.995 | unsp | PVX_003805 | 939 S | AVNPSGEEQ | 0.996 | unsp | PVX_003805 | 387 S | PAKASDEES | 0.996 | unsp | PVX_003805 | 392 S | DEESSQEEL | 0.997 | unsp |