

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_0518700 | SP | 0.018137 | 0.981422 | 0.000441 | CS pos: 24-25. ILA-IP. Pr: 0.8342 |
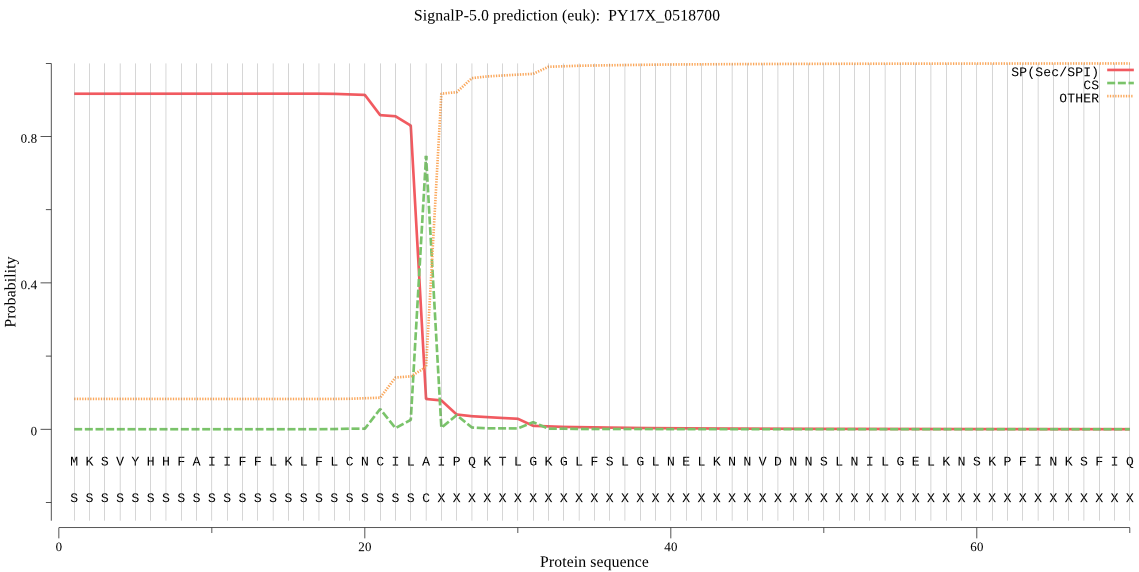
MKSVYHHFAIIFFLKLFLCNCILAIPQKTLGKGLFSLGLNELKNNVDNNSLNILGELKNS KPFINKSFIQINEKKDNVLLLKLYKQNIASDKLSTYYGKIAIGDNSENIFNVLFDTGSTE FWVPFKTCKFTKNNIHNKYERTQSFKYKYDNKGLPSVLEINYLSGKLVGFDGYDTVYLGP GFAIPHTNIAFATSIDIPVLEKFKWDGIIGLGFENEDSQKRGIKPFLDHLKDEKILTDKN YKNMFGYYITNTGGYITLGGIDNRFKRSPDEKIIWSPVSTEMGFWTIDILGIRKEKQPYM NERKDEVIVKYEGFHDGSNRSIVDTGTFLIYAPKKTMENYLNDLTINSCEDKHKLPYIIF QIKSKEIESIKGLSVIELVLSPNDYVIEYIDEVNSTKECIIGIQSDEDNINGWTLGQVFL KSYYTIFDKDNLQIGFVRNKQTLNDETYLNESFLRVNKKRNKKKSYNGPLKQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0518700 | 374 S | IKGLSVIEL | 0.991 | unsp | PY17X_0518700 | 374 S | IKGLSVIEL | 0.991 | unsp | PY17X_0518700 | 374 S | IKGLSVIEL | 0.991 | unsp | PY17X_0518700 | 381 S | ELVLSPNDY | 0.994 | unsp | PY17X_0518700 | 395 S | DEVNSTKEC | 0.994 | unsp | PY17X_0518700 | 405 S | IGIQSDEDN | 0.993 | unsp | PY17X_0518700 | 218 S | ENEDSQKRG | 0.992 | unsp | PY17X_0518700 | 279 S | WSPVSTEMG | 0.993 | unsp |