

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
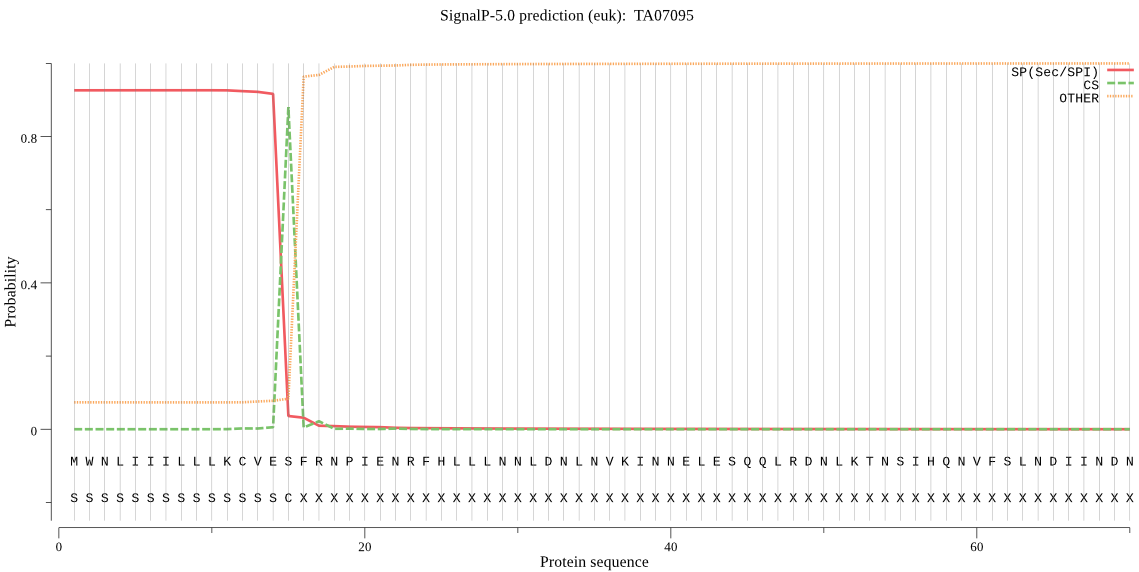
| TA07095 | SP | 0.031095 | 0.968717 | 0.000188 | CS pos: 15-16. VES-FR. Pr: 0.9639 |
MWNLIIILLLKCVESFRNPIENRFHLLLNNLDNLNVKINNELESQQLRDNLKTNSIHQNV FSLNDIINDNHNNSHDFSMCMITNTGHSYNHSPGLRYGDWSHQPAMNIKHDYKLMMMDNG GHVFNSQDYTDKAWEAITSLTEIANEFDSSYVEGDMLLYSLINDDNVLKVLNNLGLNVDN LKKELENHLKKQIRMSGSFGDRKILGRILENVLNISKRYKSEFGVSSITVLAHTATNPPK GSYIVPLDKYISVEHLLLGLAAEDTKFFRPYLTKYKITLEKLKDTVLSIRGKRKITTRNT ENSYKLLNKFSKDLTDMARNGKLDPVIGRDNEIRRTIEILSRRTKNNPVLLGDPGVGKTA IAEGLANRIVSGDVPDSLKNRKVISLDIAAIVAGTMYRGEFEERLKEILNEIENSQGEIV MFIDEIHTLVGAGESQGSLDAGNILKPMLARGELRCIGATTLQEYRQKIEKDKALERRFQ PIYIDEPNIEETINILRGLKERYEVHHGVRILDSTLIQAVLLSNRYITDRYLPDKAIDLI DEAAAKLKIQLSSKPLQLDIIERKLLQLEMEKISIINDNDNIGILSKNEKENLKPNDKLR LQNIDNLVNELTKQKDELNEMWLKEKSLVDNIRNIKERIDIVKIEIDKAERDFDLNHAAE LRFETLPDLENQLKTNINNYENYIKQVRYGVRSTATNTTNPPEGVLYSSFSYITTYIVME TGGNILLRDTVTKEDIANVVSKWTGVVRGSYILVVGSSVTVVIPFTWILSYRLTKHRNVV IGIPLNKLIKSQKEKILQLNEELHKRIIGQQEAIDAVVNAVQRSRVGMNDPKKPIAALMF LGPTGVGKTELSKAIAEQLFDSGITLRCLLPRPPTTTKDAIIRFDMSEYMEKHSVSKLVG APPGYIGYEQGGLLTEAIRRKPYSILLFDEMYLHYGACSSPTGVLSYILKSTTHSDVYNI LLQVLDDGRLTDSLGRKVNFTNSLIIFTSNLGSQNILELARFPEKRNEMKNKVMTSVRQN FSPEFLNRIDEFIVFDSLTKIGTLRCSVTVLGHTVTNTPKRELKKIVNMEMMKLSNRLAE KNIKLSIDDAAMSHIADIGYDPAYGARPLKRTIQKQIESPIAVGILSDQYKEHDNLHISY KDGKLTILPL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA07095 | 876 T | RPPTTTKDA | 0.992 | unsp | TA07095 | 876 T | RPPTTTKDA | 0.992 | unsp | TA07095 | 876 T | RPPTTTKDA | 0.992 | unsp | TA07095 | 1086 S | NIKLSIDDA | 0.993 | unsp | TA07095 | 1139 S | NLHISYKDG | 0.997 | unsp | TA07095 | 196 S | QIRMSGSFG | 0.996 | unsp | TA07095 | 377 S | DVPDSLKNR | 0.993 | unsp |