

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_202910 | SP | 0.015059 | 0.984418 | 0.000523 | CS pos: 22-23. ASA-EL. Pr: 0.7677 |
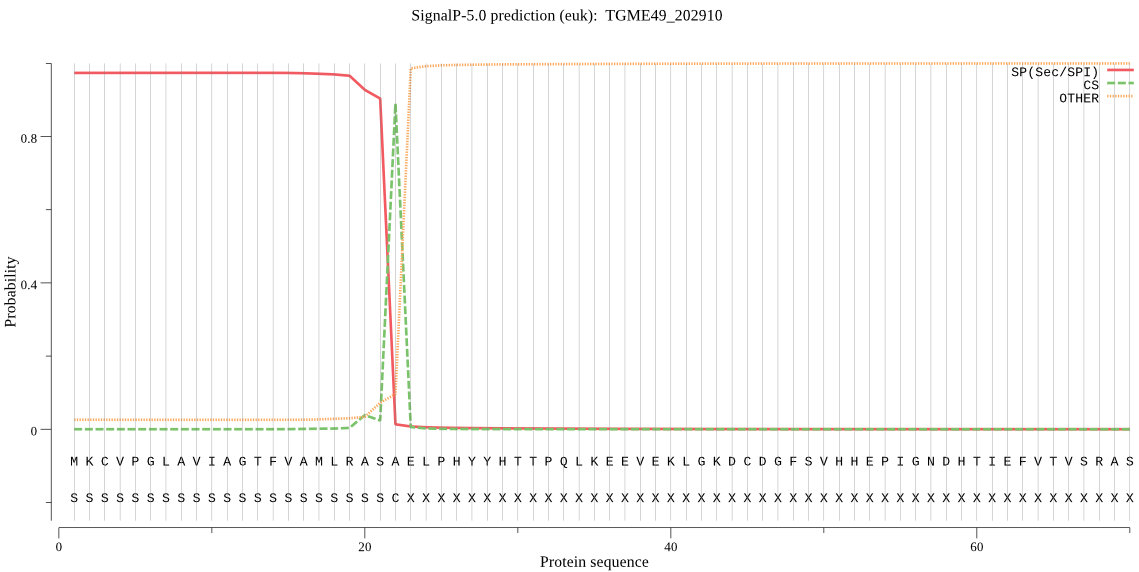
MKCVPGLAVIAGTFVAMLRASAELPHYYHTTPQLKEEVEKLGKDCDGFSVHHEPIGNDHT IEFVTVSRASGTRDRFFLLAGEHSRELISAESALHFLQVLCERETKLAEKAGKVLQHTNI QVVVNGNPVSRAKVEAGDFCLRGNENNVDLNRNWGDHWNSAAANAETSPGNKAFSEPETQ AFAKAVTSFKPHIFLSVHSGTLGMYMPWAFSAEEPARNEQAMESVLKEVDEKFCRCPYGA AGKEVGYQSSGTCLDFVYDKLKTPLSFAVEVCNLPRASDFHFKSNIVRFTPTPTDKLSSI KHGRRKSSFWIPTQTSVD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_202910 | 299 S | DKLSSIKHG | 0.996 | unsp | TGME49_202910 | 299 S | DKLSSIKHG | 0.996 | unsp | TGME49_202910 | 299 S | DKLSSIKHG | 0.996 | unsp | TGME49_202910 | 70 S | VSRASGTRD | 0.998 | unsp | TGME49_202910 | 175 S | NKAFSEPET | 0.994 | unsp |