

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
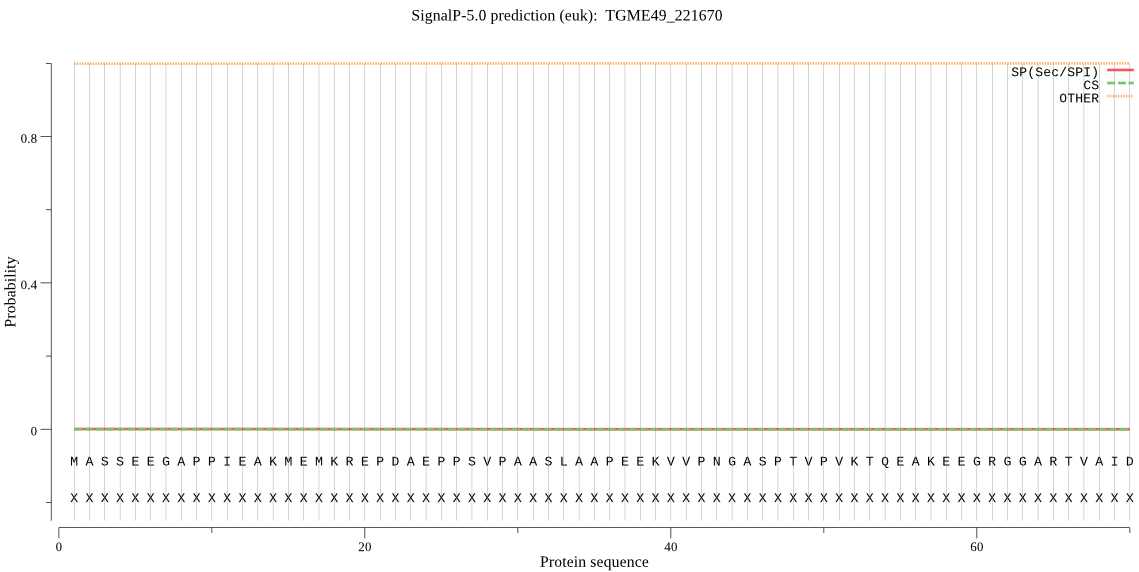
| TGME49_221670 | OTHER | 0.999989 | 0.000008 | 0.000002 |
MASSEEGAPPIEAKMEMKREPDAEPPSVPAASLAAPEEKVVPNGASPTVPVKTQEAKEEG RGGARTVAIDGGDVKEKLDRLFSYWDSCYGVEGDSWHGVDAFVILVGKASEEEEGKAEQM QMWLTGFQFPETLFVFTRTGEWWVLTSPKKLEHLRQVESCREGIFLLSRADGLPEAMEKI HQAIGRAAAAAGKNADEASIGCLQQSASLSQGGGFGQQVSDSLLRKFTESNRSKFVDDGI VATMAVHTRVEIENIRSASVVCVAMVKTQIVNRIETVLDNEQQESHAAIADLAEKLLKDG KQIEKLREKRNIDPSEVDLLYSNVQSGDVFDLRASAQPTNANLSQSEGSIIVSLGVKYKE LCAAVARTLLLNGTKEQKEVYSFTFELLNYVISLLKPGASFSSIYADARAFVEEKKPALA DHLLKMVGHCMGLEYRSNSLVLNAKNSKSVVERGMVFNISVGFSHLTTAKGKNYAIWLAD TVLLPKEEGAAPVVLTDGTSKVLRHVSYELEDAEEEPAEKKDKASADDKKRAKEETTKGA DDPERQKKKSVATKQKDSSRVKEKEKETRKAATTGGAISATILNNAESVILKDRLRRRTG SQAATAQQEAEERDERQRQLRKKKSEQLRLRFEEEKDGGGLERKKKEGKKMEDIKCFSGP EGFPRDVKANKLYVDFKSESLLVPIHGSHLPFHLSTVKNVTCSEAQGDSSGSSSVSGKNK APFYVLRINFQVPGSQTLTLKGEENPLPDLSGKPDTVFIKELMFKSEDGRHLQTIFRTIK EQLKRVKQKALEDDVAGEVTEQDKLILNRSGRRVLLKDLMIRPNIAPGMRKLIGALEAHT NGLRFTVNTRGQIDQVDITYSNIKHAMFQPCERELIVLIHFHLKSAIMVGKKRTQDVQFY TEAGTQTDDLDNRRNRSFHDPDETQDEMRERELKRRLNNEFKRFVQQVEDIAKVEFDLPY RELRFTGVPMKSNVEILPTANCLVHLIEWPPFVLPLEDIELVSFERVAHGLRNFDVIFVF QDYTKPVKRIDLVPIEFLDNLKRWLNELEIVWYEGKQNLNWNAILKQIREDPHGFVEAGG FEMFLGDDSVSGEEGDTDEDDDDEEYAESGSESEYNERSGEEEEDGEEGSSEEDSSDSDD DESLADESDEDEEYNDVSSDEEEGLSWDELEERAKKEDRKRRTDDSDDNEDRRKKKRK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_221670 | 344 S | NANLSQSEG | 0.996 | unsp | TGME49_221670 | 344 S | NANLSQSEG | 0.996 | unsp | TGME49_221670 | 344 S | NANLSQSEG | 0.996 | unsp | TGME49_221670 | 525 S | KDKASADDK | 0.996 | unsp | TGME49_221670 | 716 S | SSSVSGKNK | 0.995 | unsp | TGME49_221670 | 917 S | RRNRSFHDP | 0.996 | unsp | TGME49_221670 | 1091 S | DDSVSGEEG | 0.998 | unsp | TGME49_221670 | 1113 S | SGSESEYNE | 0.99 | unsp | TGME49_221670 | 1119 S | YNERSGEEE | 0.996 | unsp | TGME49_221670 | 1130 S | GEEGSSEED | 0.99 | unsp | TGME49_221670 | 1131 S | EEGSSEEDS | 0.998 | unsp | TGME49_221670 | 1136 S | EEDSSDSDD | 0.997 | unsp | TGME49_221670 | 1138 S | DSSDSDDDE | 0.997 | unsp | TGME49_221670 | 1143 S | DDDESLADE | 0.991 | unsp | TGME49_221670 | 1148 S | LADESDEDE | 0.997 | unsp | TGME49_221670 | 1159 S | NDVSSDEEE | 0.996 | unsp | TGME49_221670 | 1166 S | EEGLSWDEL | 0.994 | unsp | TGME49_221670 | 1186 S | RTDDSDDNE | 0.991 | unsp | TGME49_221670 | 110 S | VGKASEEEE | 0.992 | unsp | TGME49_221670 | 315 S | NIDPSEVDL | 0.993 | unsp |