

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_254010 | SP | 0.469678 | 0.508444 | 0.021878 | CS pos: 48-49. VEA-TG. Pr: 0.8922 |
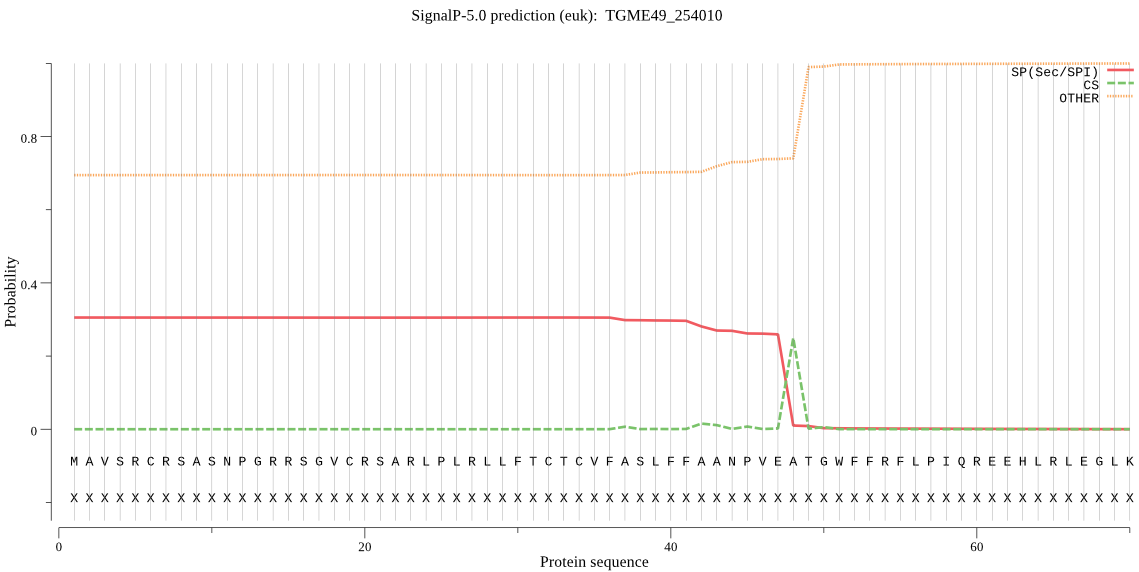
MAVSRCRSASNPGRRSGVCRSARLPLRLLFTCTCVFASLFFAANPVEATGWFFRFLPIQR EEHLRLEGLKASGTPPQAGRHTREEESARARGEEKERDREASASEGGEKRRDGGDGGLRV IQEKETGRGSDERSEEGRSSSNLLPGFLRRFLPSPSFQSGQVNLSTDQERRLETDTDTNE AGDRERREQWGGEGSRSREADRGELEVQEGWFLQPLDHGNPLVFNRLGHDAWRQKFYRAK RKRRSSKEEESTEVAGHAKTERQNDSLTDDAIRPIFVYIGGEGPLSSLEVRQGLLAEMGD IFGASLYALEHRYYGDSHPRPDSSVVNLQWLTSHQALGDLAAFVAHVKQQEAEEHPQDLA PEDVPVVVFGCSYPGSLAAYARAKYPASILGAVSSSSPVEASALFQAFDRVVQRVLPAAC TAKVKAATAVVERRLFSGEEEAVKVAAKFGCGADVPMKTHDQRVALLYVIADAIAESVQY NRQPTRPWIEEVCACFSETASEREETHDNKGDKREKRDNEEDLVNALAKAVQLMLAKLKM TCKDSNLLQLTDTRLGPQASASARLWTWQSCAEYGYWQVAYKDSVRSHLIDLDWHMRMCN ALFPLPSGSKFSTDVVAETNVWSGDKLVAGVGAATNIHFTNGENDPWAPLSVTEVSPVVV DRQGLSSFTIQDGSHCNDFYAYGGTEPVAVTEAKARIQNAIRAWLEDFRERREQQKRKVD PPLTKTFSATSVGGDSEL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_254010 | 197 S | EGSRSREAD | 0.995 | unsp | TGME49_254010 | 197 S | EGSRSREAD | 0.995 | unsp | TGME49_254010 | 197 S | EGSRSREAD | 0.995 | unsp | TGME49_254010 | 245 S | RKRRSSKEE | 0.998 | unsp | TGME49_254010 | 246 S | KRRSSKEEE | 0.998 | unsp | TGME49_254010 | 397 S | VSSSSPVEA | 0.997 | unsp | TGME49_254010 | 437 S | RRLFSGEEE | 0.997 | unsp | TGME49_254010 | 501 S | SETASEREE | 0.995 | unsp | TGME49_254010 | 102 S | DREASASEG | 0.997 | unsp | TGME49_254010 | 130 S | TGRGSDERS | 0.998 | unsp |