

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
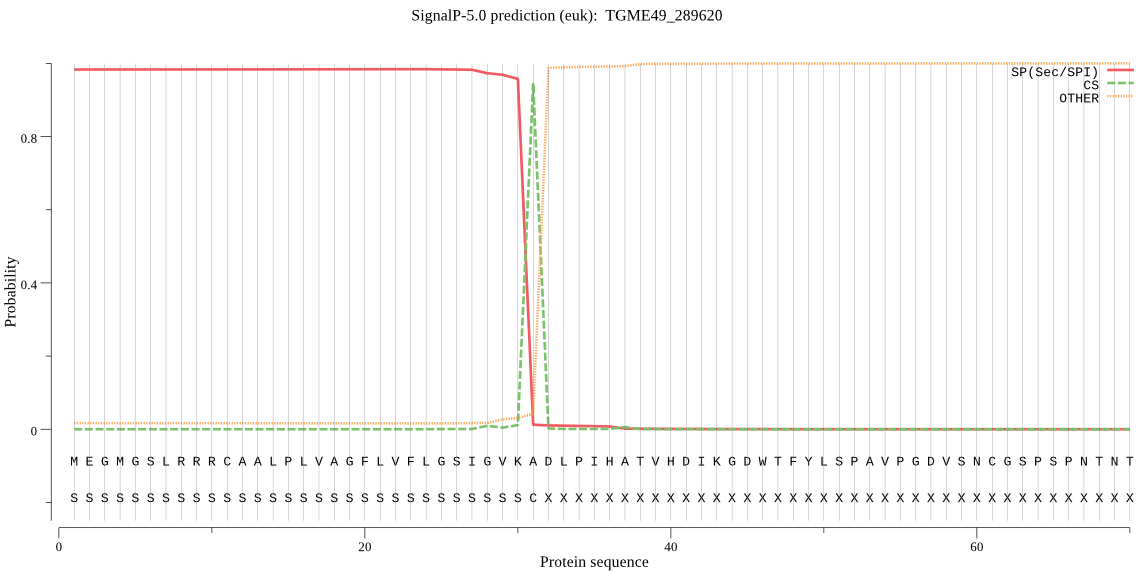
| TGME49_289620 | SP | 0.004204 | 0.995747 | 0.000049 | CS pos: 31-32. VKA-DL. Pr: 0.9411 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_289620 | 320 S | STDISTADG | 0.996 | unsp | TGME49_289620 | 320 S | STDISTADG | 0.996 | unsp | TGME49_289620 | 320 S | STDISTADG | 0.996 | unsp | TGME49_289620 | 420 S | PKDFSWSDP | 0.995 | unsp | TGME49_289620 | 97 S | ELRVSLTDI | 0.998 | unsp | TGME49_289620 | 271 S | SPASSWRAG | 0.994 | unsp |