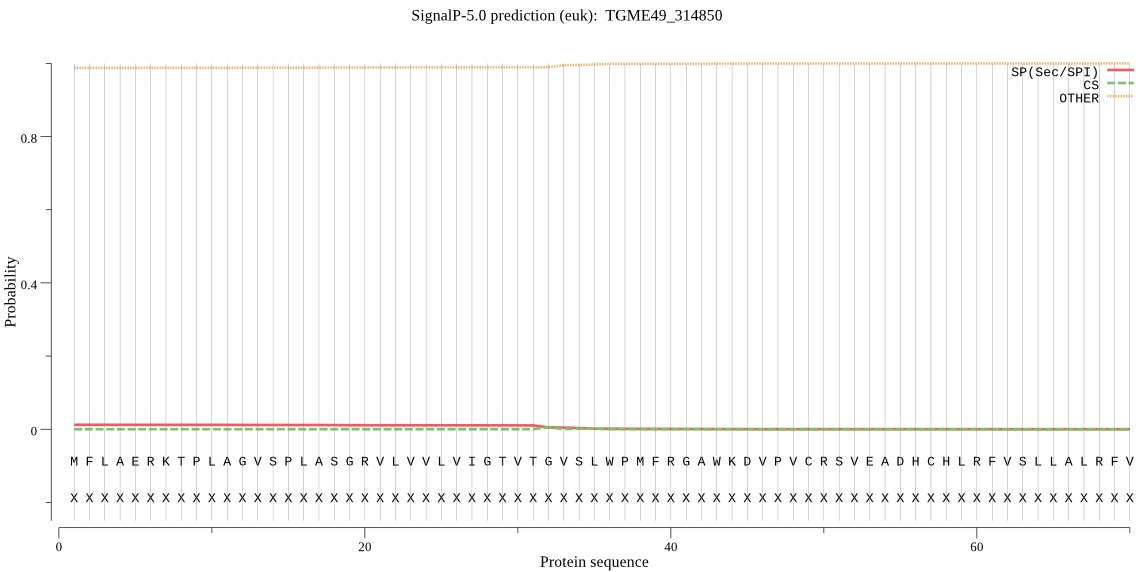
Fasta :-
MFLAERKTPLAGVSPLASGRVLVVLVIGTVTGVSLWPMFRGAWKDVPVCRSVEADHCHLR
FVSLLALRFVSRLLSLLSAYCVFCPLVVSASSSSSPLSRGAPSSRKAPLVFSTEISSRAS
NPASDLFGGPASDAQARGCLRRSIPEAVALRAKSASPLFSLFSRLEMALVDTLALVPSLA
ALASPSPPSPSPGRPGVSVPPRPLLKRSSSLVVLGLLVHLLSGSPGVAPVDVLRSFDRLF
LAEAAQLLRDDVEAQSSTNRRKSAVADPDDWETQRREWRYPQARGPAETGSSVPPGDSFG
AGAQRQRENGDPVHTHSLRAPFVLEGNEIPAPRSDHRFRRFVRLANGMEILILVDPLQLR
SAPSHVSLSVAGGFLHDPAHLPGLATLSGHLIFNNYVPVSSALSAASLAGGVGSEQKSLG
RRAESRNRRGEGGRDPEQEQGREEPSSDSESGGMFPSLDAFLFAHDAAEPLHFSTGEFAS
ALSYSLPTPALAASLAELKHILLHPNITDASVGVGVVQLLQQQQKQVQKRKSEPYWRKAS
ILKNHVFNPAHKTSRFHVGSTDLIFLLLHHQQEAAARTAVEGSGRGAGSGREAKDPVFER
VSLPMFQHVNVWLKNEDIPSLVWEWHKRFYVANAFKLVVSTPGSQRDLDRLTDQIQRLFD
PLPSNPALEHAFTCKADAETPFFASAQEAEAFAHQELEASMRATAHSRRGNKRDRAEKCQ
RRGTGRASEGPEESAGVLEEGDRNRDSSGTFFQPSRVAQVACGPARERPSQGEGDRETRR
GPGETAAGRNGSEERRAETSDSRCGSSAEATGQTSCAVADDSAKTKETDDGARKRDDPTE
VPVARHCDLRKEILLEPATENEHSVDFVFVFEKPHPKWTVFDAKYVVDVLGSEGPGSLVA
HLRKQGLAESIDVEAEESRCYSTLRASVELKDFNMNHTAIMLIGSSLFSYLRFIRESVLD
RAFVESRMRMYRLAFDAHVPPLPPRPPLPGALGTIPALLPSSLLQSSLPSSSPGQAGTNF
SPQEEVAYLAADLQEILSPARVFTEQWLLEDVGKQSLEHYLDQLTPDNLILMISSPLVVP
LCTLVSSFFGIPFAINPLDATQDAAWSALVSLPPERALLLARRTGFSLPPASLFVPETPA
HVDHHLGDNAVDRFGQARGGGGRAADEPEAEAGGAETVEPNSRGPARSGADASSLSEEEE
RVLFSSRQVPRLLAFEDGEESTGLSCSGHCELFHLPYRTPEQNPPRASVTLLLNYAVPSL
SLLREDEQATQAETKMGGAKGRTGGEPPLRDTAISPTRYREAVVSLMGLYVATVKEMLHE
VTVYASMAEIAFSIEADQYMSTVEIHVEGVSAVIPVLVELIVKGLMAGSPGSPVAGRAPP
EGDAGRESPATAVPPLPQVFACRNPWKDACGDFLDLGAFERVKQEKIDELREGAHDLPSC
LAEVVFTTLLRDRDWGGRHSKLDVIRHARLRDVEDAGALLKGKLFVQALVAGDMSETAAK
RLMSNVSSRLQLSGKPAVEELSWEPVLSMKKMNFPSFVERQIRHRGGRALPAVPRYRWRL
AYGSWVESNVCSPFSLSTSPSFELVDEDRLDARQRSESTGKASKAKIDQERGTPKTANGP
AKEAPLTHPDGLHGPLLGMSALRIELGVLTDKQLAIAAVLELLLAYPMYRHVCALDGACH
SLSFSLRQEPAGVSYFLLDLRTFEVPACVATERAEAWFFTQAGVFSGMFPTRGDASHVSS
WTLNSASPGSAESPASQADVALYPSDGETVLLRRDFLKKQYAHNPAIRSVPRKASEAREA
RGKPEQETSFQERPSGSCAGAACPEAVGRRASFGASEGGTGDAGSAAVSPSGQDDSSEGE
AGEREEREEREHAASCSESTHGADRLPRGGVSPDLSGSSCTASRESVDDGSRSGAPGLAS
AFDPRQHRAFEEPGFFVFLDRDRFQKAKDAAEASFLMLYDWEADHVQRRRRREAERSASG
REEDGNDAKRDAKRTENPETEEGEADTFLGRPFIPADIQMFVHEIRKRTYVFARREQVAR
HIRSLTLEDAREFFLNSVIVAPWLVVELSNLLVPSSSNNFDGFSEPHATSGVGGMMTSGP
EACSSSDSFYAERGMPDMTTLDMRSSSNPLWRLRNSWVDVESINGFREATMNAYTLGIPE
LYRRQGRVSKQQSA