

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| YEL060C | SP | 0.443747 | 0.555318 | 0.000935 | CS pos: 18-19. ISA-AL. Pr: 0.4988 |
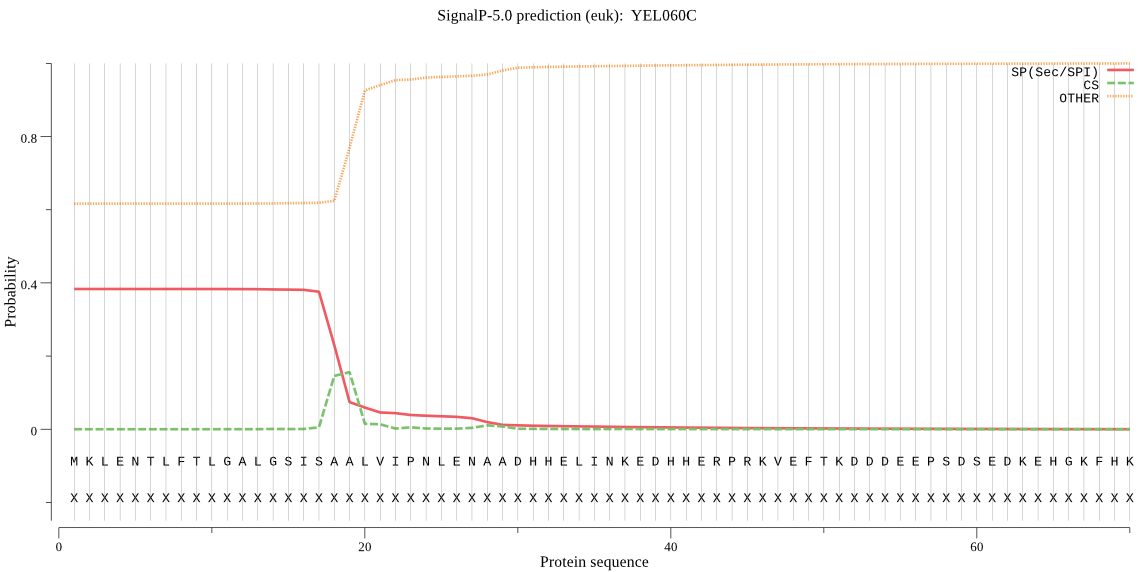
MKLENTLFTLGALGSISAALVIPNLENAADHHELINKEDHHERPRKVEFTKDDDEEPSDS EDKEHGKFHKKGRKGQDKESPEFNGKRASGSHGSAHEGGKGMKPKHESSNDDDNDDKKKK PHHKGGCHENKVEEKKMKGKKVKGKKHHEKTLEKGRHHNRLAPLVSTAQFNPDAISKIIP NRYIIVFKRGAPQEEIDFHKENVQQAQLQSVENLSAEDAFFISTKDTSLSTSEAGGIQDS FNIDNLFSGYIGYFTQEIVDLIRQNPLVDFVERDSIVEATEFDTQNSAPWGLARISHRER LNLGSFNKYLYDDDAGRGVTSYVIDTGVNINHKDFEKRAIWGKTIPLNDEDLDGNGHGTH CAGTIASKHYGVAKNANVVAVKVLRSNGSGTMSDVVKGVEYAAKAHQKEAQEKKKGFKGS TANMSLGGGKSPALDLAVNAAVEVGIHFAVAAGNENQDACNTSPASADKAITVGASTLSD DRAYFSNWGKCVDVFAPGLNILSTYIGSDDATATLSGTSMASPHVAGLLTYFLSLQPGSD SEFFELGQDSLTPQQLKKKLIHYSTKDILFDIPEDTPNVLIYNGGGQDLSAFWNDTKKSH SSGFKQELNMDEFIGSKTDLIFDQVRDILDKLNII
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YEL060C | 108 S | PKHESSNDD | 0.991 | unsp | YEL060C | 108 S | PKHESSNDD | 0.991 | unsp | YEL060C | 108 S | PKHESSNDD | 0.991 | unsp | YEL060C | 215 S | VENLSAEDA | 0.997 | unsp | YEL060C | 223 S | AFFISTKDT | 0.995 | unsp | YEL060C | 230 S | DTSLSTSEA | 0.994 | unsp | YEL060C | 275 S | VERDSIVEA | 0.997 | unsp | YEL060C | 296 S | LARISHRER | 0.995 | unsp | YEL060C | 58 S | DEEPSDSED | 0.994 | unsp | YEL060C | 80 S | QDKESPEFN | 0.996 | unsp |